Welcome to RSAT Training material
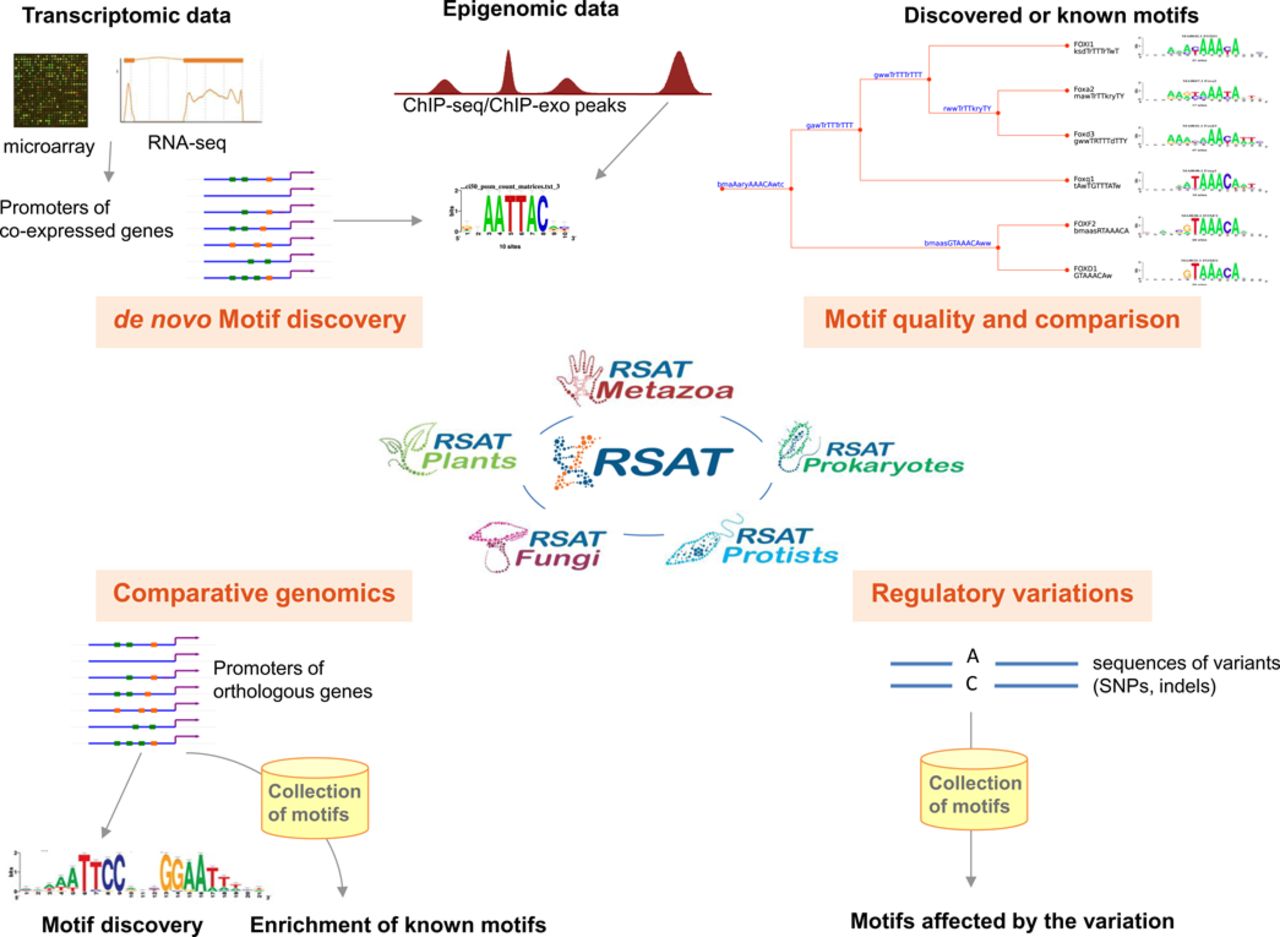
The Regulatory Sequence Analysis Tools (RSAT) comprises modular computer programs specifically designed for the detection of regulatory signals in non-coding sequences. RSAT servers have been up and running since 1997. The project was initiated by Jacques van Helden, and is now pursued by the RSAT team.
A dedicated server for learning
To learn how to use RSAT, we recommend you to use the dedicated server http://teaching.rsat.eu/.
Although tutorials and protocols used to indicate other URLs (like http://rsat.ulb.ac.be), we now recommend to use http://teaching.rsat.eu/.
Where to start ?
- To get an overview of the tools, you may read the latest description in NAR web server issue:
Medina-Rivera A, Defrance M, Sand O, Herrmann C, Castro-Mondragon J, Delerce J, Jaeger S, Blanchet C, Vincens P, Caron C, Staines DM, Contreras-Moreira B, Artufel M, Charbonnier-Khamvongsa L, Hernandez C, Thieffry D, Thomas-Chollier M, van Helden J (2015) RSAT 2015: Regulatory Sequence Analysis Tools . Nucleic Acids Res. 2015 (Web Server issue) in press.
-
Several protocols have been published to guide new users. Some of the content might be outdated, but the theory and general principles are still valid
-
ChIP-seq analysis Thomas-Chollier M, Darbo E, Herrmann C, Defrance M, Thieffry D, van Helden J. (2012). A complete workflow for the analysis of full-size ChIP-seq (and similar) data sets using peak-motifs. Nat Protoc 7(8): 1551-1568.
-
Scanning sequences with motifs Turatsinze, J.V., Thomas-Chollier, M., Defrance, M. and van Helden, J. (2008) Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat Protoc, 3, 1578-1588.</a>
-
Motif discovery Defrance, M., Janky, R., Sand, O. and van Helden, J. (2008) Using RSAT oligo-analysis and dyad-analysis tools to discover regulatory signals in nucleic sequences. Nat Protoc, 3, 1589-1603.
-
Web services access (advanced usage for bioinformaticians) Sand, O., Thomas-Chollier, M., Vervisch, E. and van Helden, J. (2008) Analyzing multiple data sets by interconnecting RSAT programs via SOAP Web services-an example with ChIP-chip data. Nat Protoc, 3,1604-1615.
-
Hands-on Tutorials
-
Tutorials of individual tools on the RSAT Web site: http://teaching.rsat.eu/tutorials.php
-
ECCB 2014 tutorial (covers access via Web site, command-line and Web services): Analysis of Cis-Regulatory Motifs from High-Throughput Sequence Sets.
Complete course on analysis of regulatory sequences
- Course by Jacques van Helden : http://rsa-tools.github.io/course/
Which tool to choose ?
The home page of each RSAT server has a section to help finding the appropriate program, based on the type of data and question. Only main tasks are supported, as this is meant to new users.
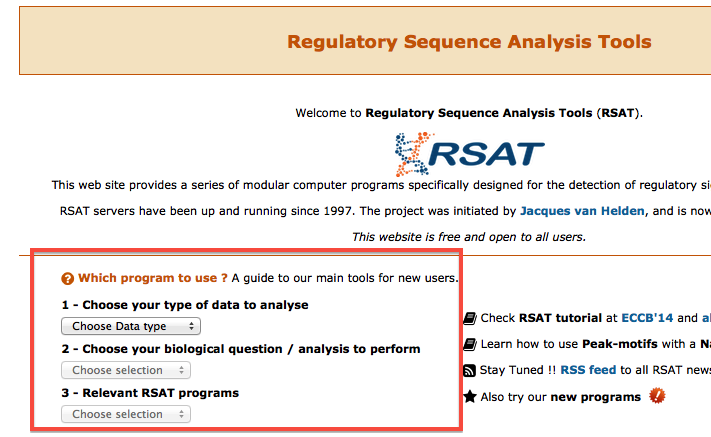
Also check the map of the tools (version 2012):

Even more resources…
-
Workshop: ChIP-Seq Data Analysis, in the context of Ecole de bioinformatique AVIESAN 2014, Station Biologique, Roscoff, France.
-
Course by Jacques van Helden : Statistics for bioinformatics
-
ALLBIO training workshop 2014 Analysing thousands of bacterial genomes: gene annotation, metabolism and regulation
Contact and Questions
Check out the RSAT forum to see questions of users or to contact the RSAT team.
