One-to-n alignments
Command: compare-matrices -v 1 -mode matches -format1 transfac -file1 $RSAT/public_html/tmp/wwwrun/2014/09/03/peak-motifs.2014-09-03.094519_2014-09-03.094519_gM9GBj/results/discovered_motifs/oligos_6nt_mkv4_m1/peak-motifs_oligos_6nt_mkv4_m1.tf -format2 tf -file2 $RSAT/public_html/data/motif_databases/JASPAR/jaspar_core_vertebrates_2013-11.tf -mode matches -DR -uth offset_rank 1 -lth w 5 -lth Wr 0.3 -lth cor 0.75 -lth Ncor 0.4 -return matrix_name,matrix_id,cor,Ncor,logoDP,NIcor,NsEucl,SSD,NSW,match_rank,width,strand,offset,consensus,alignments_1ton -sort Ncor -o $RSAT/public_html/tmp/wwwrun/2014/09/03/peak-motifs.2014-09-03.094519_2014-09-03.094519_gM9GBj/results/discovered_motifs/oligos_6nt_mkv4_m1/peak-motifs_oligos_6nt_mkv4_m1_vs_db_jaspar_core_vertebrates
One-to-n matrix alignment; reference matrix: oligos_6nt_mkv4_m1_shift1 ; 6 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| oligos_6nt_mkv4_m1_shift1 (oligos_6nt_mkv4_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_6nt_mkv4_m1; m=0 (reference); ncol1=15; shift=1; ncol=16; -dkATTGTGCAATCwt
; Alignment reference
a 0 1020 725 2836 11 6 122 129 5 244 3304 3306 61 474 1385 763
c 0 526 533 95 7 0 18 37 1 2943 18 4 304 2273 689 753
g 0 848 851 351 11 35 3080 63 3310 7 1 6 79 297 338 629
t 0 930 1215 42 3295 3283 104 3095 8 130 1 8 2880 280 912 1179
|
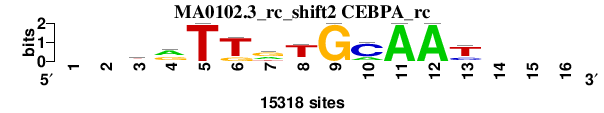
| MA0102.3_rc_shift2 (CEBPA_rc) |
 |
0.948 |
0.695 |
4.203 |
0.060 |
0.946 |
0.706 |
0.968 |
1 |
1 |
4 |
4 |
1 |
1 |
1 |
1.857 |
1 |
; oligos_6nt_mkv4_m1 versus MA0102.3_rc (CEBPA_rc); m=1/5; ncol2=11; w=11; offset=1; strand=R; shift=2; score= 1.8571; --krTTrTGCAAt---
; cor=0.948; Ncor=0.695; logoDP=4.203; NIcor=0.060; NsEucl=0.946; SSD=0.706; NSW=0.968; rcor=1; rNcor=1; rlogoDP=4; rNIcor=4; rNsEucl=1; rSSD=1; rNSW=1; rank_mean=1.857; match_rank=1
a 0 0 3661 7826 0 0 5004 1022 0 3309 15318 15318 0 0 0 0
c 0 0 890 1804 0 0 0 1066 0 11304 0 0 3635 0 0 0
g 0 0 4963 5688 0 3092 7654 1274 15318 0 0 0 1332 0 0 0
t 0 0 5804 0 15318 12226 2660 11956 0 705 0 0 10351 0 0 0
|
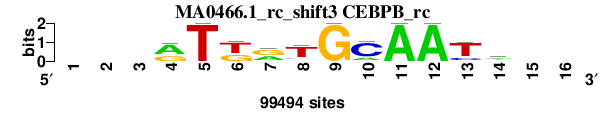
| MA0466.1_rc_shift3 (CEBPB_rc) |
 |
0.924 |
0.678 |
2.820 |
-0.039 |
0.935 |
1.012 |
0.954 |
2 |
2 |
5 |
5 |
2 |
2 |
2 |
2.857 |
2 |
; oligos_6nt_mkv4_m1 versus MA0466.1_rc (CEBPB_rc); m=2/5; ncol2=11; w=11; offset=2; strand=R; shift=3; score= 2.8571; ---rTkrTGCAATm--
; cor=0.924; Ncor=0.678; logoDP=2.820; NIcor=-0.039; NsEucl=0.935; SSD=1.012; NSW=0.954; rcor=2; rNcor=2; rlogoDP=5; rNIcor=5; rNsEucl=2; rSSD=2; rNSW=2; rank_mean=2.857; match_rank=2
a 0 0 0 61413 0 0 38886 9286 0 19407 99494 99494 0 42845 0 0
c 0 0 0 2043 0 0 0 10015 0 75531 0 0 18428 33617 0 0
g 0 0 0 36038 0 39343 51954 5478 99494 0 0 0 5868 10026 0 0
t 0 0 0 0 99494 60151 8654 74715 0 4556 0 0 75198 13006 0 0
|
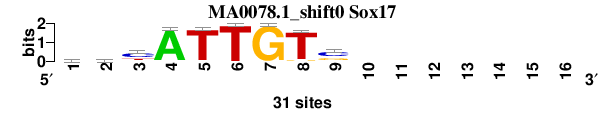
| MA0078.1_shift0 (Sox17) |
 |
0.861 |
0.431 |
6.882 |
0.426 |
0.908 |
1.089 |
0.932 |
3 |
5 |
1 |
3 |
4 |
3 |
3 |
3.143 |
3 |
; oligos_6nt_mkv4_m1 versus MA0078.1 (Sox17); m=3/5; ncol2=9; w=8; offset=-1; strand=D; shift=0; score= 3.1429; yhyATTGTs-------
; cor=0.861; Ncor=0.431; logoDP=6.882; NIcor=0.426; NsEucl=0.908; SSD=1.089; NSW=0.932; rcor=3; rNcor=5; rlogoDP=1; rNIcor=3; rNsEucl=4; rSSD=3; rNSW=3; rank_mean=3.143; match_rank=3
a 7 8 3 30 0 0 0 0 0 0 0 0 0 0 0 0
c 9 8 18 0 1 0 0 0 17 0 0 0 0 0 0 0
g 6 4 1 0 0 0 31 2 10 0 0 0 0 0 0 0
t 9 11 9 1 30 31 0 29 4 0 0 0 0 0 0 0
|
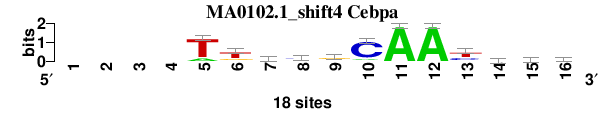
| MA0102.1_shift4 (Cebpa) |
 |
0.811 |
0.649 |
6.234 |
0.631 |
0.913 |
2.195 |
0.909 |
5 |
3 |
2 |
1 |
3 |
5 |
4 |
3.286 |
4 |
; oligos_6nt_mkv4_m1 versus MA0102.1 (Cebpa); m=4/5; ncol2=12; w=12; offset=3; strand=D; shift=4; score= 3.2857; ----TtkygCAAyvyb
; cor=0.811; Ncor=0.649; logoDP=6.234; NIcor=0.631; NsEucl=0.913; SSD=2.195; NSW=0.909; rcor=5; rNcor=3; rlogoDP=2; rNIcor=1; rNsEucl=3; rSSD=5; rNSW=4; rank_mean=3.286; match_rank=4
a 0 0 0 0 3 1 4 2 4 2 18 18 0 5 2 1
c 0 0 0 0 0 1 1 9 2 15 0 0 6 6 5 6
g 0 0 0 0 0 4 6 2 10 0 0 0 2 5 3 5
t 0 0 0 0 15 12 7 5 2 1 0 0 10 2 8 6
|
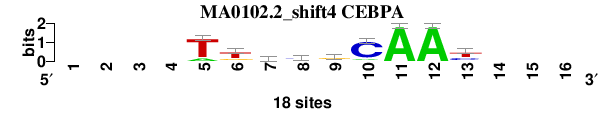
| MA0102.2_shift4 (CEBPA) |
 |
0.839 |
0.503 |
6.221 |
0.480 |
0.894 |
1.836 |
0.898 |
4 |
4 |
3 |
2 |
5 |
4 |
5 |
3.857 |
5 |
; oligos_6nt_mkv4_m1 versus MA0102.2 (CEBPA); m=5/5; ncol2=9; w=9; offset=3; strand=D; shift=4; score= 3.8571; ----TtkygCAAy---
; cor=0.839; Ncor=0.503; logoDP=6.221; NIcor=0.480; NsEucl=0.894; SSD=1.836; NSW=0.898; rcor=4; rNcor=4; rlogoDP=3; rNIcor=2; rNsEucl=5; rSSD=4; rNSW=5; rank_mean=3.857; match_rank=5
a 0 0 0 0 3 1 4 2 4 2 18 18 0 0 0 0
c 0 0 0 0 0 1 1 9 2 15 0 0 6 0 0 0
g 0 0 0 0 0 4 6 2 10 0 0 0 2 0 0 0
t 0 0 0 0 15 12 7 5 2 1 0 0 10 0 0 0
|





