One-to-n alignments
Command: compare-matrices -v 1 -mode matches -format1 transfac -file1 $RSAT/public_html/tmp/wwwrun/2014/09/03/peak-motifs.2014-09-03.094519_2014-09-03.094519_gM9GBj/results/discovered_motifs/oligos_6nt_mkv4_m2/peak-motifs_oligos_6nt_mkv4_m2.tf -format2 tf -file2 $RSAT/public_html/data/motif_databases/JASPAR/jaspar_core_vertebrates_2013-11.tf -mode matches -DR -uth offset_rank 1 -lth w 5 -lth Wr 0.3 -lth cor 0.75 -lth Ncor 0.4 -return matrix_name,matrix_id,cor,Ncor,logoDP,NIcor,NsEucl,SSD,NSW,match_rank,width,strand,offset,consensus,alignments_1ton -sort Ncor -o $RSAT/public_html/tmp/wwwrun/2014/09/03/peak-motifs.2014-09-03.094519_2014-09-03.094519_gM9GBj/results/discovered_motifs/oligos_6nt_mkv4_m2/peak-motifs_oligos_6nt_mkv4_m2_vs_db_jaspar_core_vertebrates
One-to-n matrix alignment; reference matrix: oligos_6nt_mkv4_m2_shift0 ; 2 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| oligos_6nt_mkv4_m2_shift0 (oligos_6nt_mkv4_m2) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_6nt_mkv4_m2; m=0 (reference); ncol1=10; shift=0; ncol=11; waCAAAACwa-
; Alignment reference
a 3789 3782 0 9991 9991 9991 9991 0 4608 3524 0
c 1446 1847 9991 0 0 0 0 9991 2039 2075 0
g 1693 1936 0 0 0 0 0 0 531 1897 0
t 3063 2426 0 0 0 0 0 0 2813 2495 0
|
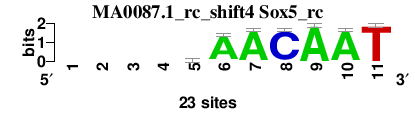
| MA0087.1_rc_shift4 (Sox5_rc) |
 |
0.767 |
0.418 |
1.994 |
0.014 |
0.855 |
1.516 |
0.874 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1.000 |
1 |
; oligos_6nt_mkv4_m2 versus MA0087.1_rc (Sox5_rc); m=1/1; ncol2=7; w=6; offset=4; strand=R; shift=4; score= 1; ----wAACAAT
; cor=0.767; Ncor=0.418; logoDP=1.994; NIcor=0.014; NsEucl=0.855; SSD=1.516; NSW=0.874; rcor=1; rNcor=1; rlogoDP=1; rNIcor=1; rNsEucl=1; rSSD=1; rNSW=1; rank_mean=1.000; match_rank=1
a 0 0 0 0 8 21 22 0 23 22 0
c 0 0 0 0 4 0 0 22 0 1 0
g 0 0 0 0 3 1 1 0 0 0 0
t 0 0 0 0 8 1 0 1 0 0 23
|

