| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| oligos_7nt_mkv5_m1_shift1 (oligos_7nt_mkv5_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_7nt_mkv5_m1; m=0 (reference); ncol1=15; shift=1; ncol=16; -wkATTGCACAATcwy
; Alignment reference
a 0 881 596 2604 4 6 132 4 2769 41 2931 2943 37 506 1027 722
c 0 545 555 127 12 0 10 2960 44 2728 35 5 195 1897 703 784
g 0 620 810 181 8 12 2652 3 37 10 2 12 67 243 299 499
t 0 926 1011 60 2948 2954 178 5 122 193 4 12 2673 326 943 967
|
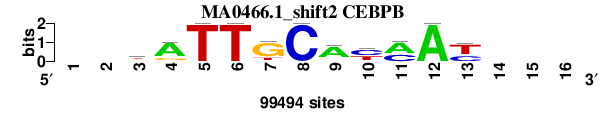
| MA0466.1_shift2 (CEBPB) |
 |
0.934 |
0.685 |
9.857 |
0.677 |
0.940 |
0.865 |
0.961 |
2 |
2 |
2 |
1 |
1 |
1 |
1 |
1.429 |
1 |
; oligos_7nt_mkv5_m1 versus MA0466.1 (CEBPB); m=1/6; ncol2=11; w=11; offset=1; strand=D; shift=2; score= 1.4286; --kATTGCAymAy---
; cor=0.934; Ncor=0.685; logoDP=9.857; NIcor=0.677; NsEucl=0.940; SSD=0.865; NSW=0.961; rcor=2; rNcor=2; rlogoDP=2; rNIcor=1; rNsEucl=1; rSSD=1; rNSW=1; rank_mean=1.429; match_rank=1
a 0 0 13006 75198 0 0 4556 0 74715 8654 60151 99494 0 0 0 0
c 0 0 10026 5868 0 0 0 99494 5478 51954 39343 0 36038 0 0 0
g 0 0 33617 18428 0 0 75531 0 10015 0 0 0 2043 0 0 0
t 0 0 42845 0 99494 99494 19407 0 9286 38886 0 0 61413 0 0 0
|
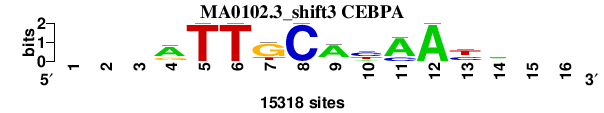
| MA0102.3_shift3 (CEBPA) |
 |
0.935 |
0.685 |
9.923 |
0.674 |
0.939 |
0.907 |
0.959 |
1 |
1 |
1 |
2 |
2 |
2 |
2 |
1.571 |
2 |
; oligos_7nt_mkv5_m1 versus MA0102.3 (CEBPA); m=2/6; ncol2=11; w=11; offset=2; strand=D; shift=3; score= 1.5714; ---aTTGCAyAAym--
; cor=0.935; Ncor=0.685; logoDP=9.923; NIcor=0.674; NsEucl=0.939; SSD=0.907; NSW=0.959; rcor=1; rNcor=1; rlogoDP=1; rNIcor=2; rNsEucl=2; rSSD=2; rNSW=2; rank_mean=1.571; match_rank=2
a 0 0 0 10351 0 0 705 0 11956 2660 12226 15318 0 5804 0 0
c 0 0 0 1332 0 0 0 15318 1274 7654 3092 0 5688 4963 0 0
g 0 0 0 3635 0 0 11304 0 1066 0 0 0 1804 890 0 0
t 0 0 0 0 15318 15318 3309 0 1022 5004 0 0 7826 3661 0 0
|
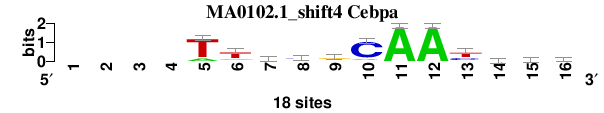
| MA0102.1_shift4 (Cebpa) |
 |
0.797 |
0.637 |
6.191 |
0.620 |
0.910 |
2.314 |
0.904 |
5 |
3 |
3 |
3 |
3 |
5 |
4 |
3.714 |
3 |
; oligos_7nt_mkv5_m1 versus MA0102.1 (Cebpa); m=3/6; ncol2=12; w=12; offset=3; strand=D; shift=4; score= 3.7143; ----TtkygCAAyvyb
; cor=0.797; Ncor=0.637; logoDP=6.191; NIcor=0.620; NsEucl=0.910; SSD=2.314; NSW=0.904; rcor=5; rNcor=3; rlogoDP=3; rNIcor=3; rNsEucl=3; rSSD=5; rNSW=4; rank_mean=3.714; match_rank=3
a 0 0 0 0 3 1 4 2 4 2 18 18 0 5 2 1
c 0 0 0 0 0 1 1 9 2 15 0 0 6 6 5 6
g 0 0 0 0 0 4 6 2 10 0 0 0 2 5 3 5
t 0 0 0 0 15 12 7 5 2 1 0 0 10 2 8 6
|
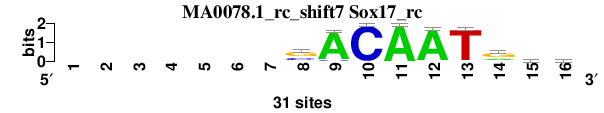
| MA0078.1_rc_shift7 (Sox17_rc) |
 |
0.816 |
0.490 |
3.080 |
0.177 |
0.904 |
1.499 |
0.917 |
4 |
6 |
4 |
4 |
4 |
3 |
3 |
4.000 |
4 |
; oligos_7nt_mkv5_m1 versus MA0078.1_rc (Sox17_rc); m=4/6; ncol2=9; w=9; offset=6; strand=R; shift=7; score= 4; -------sACAATrdr
; cor=0.816; Ncor=0.490; logoDP=3.080; NIcor=0.177; NsEucl=0.904; SSD=1.499; NSW=0.917; rcor=4; rNcor=6; rlogoDP=4; rNIcor=4; rNsEucl=4; rSSD=3; rNSW=3; rank_mean=4.000; match_rank=4
a 0 0 0 0 0 0 0 4 29 0 31 30 1 9 11 9
c 0 0 0 0 0 0 0 10 2 31 0 0 0 1 4 6
g 0 0 0 0 0 0 0 17 0 0 0 1 0 18 8 9
t 0 0 0 0 0 0 0 0 0 0 0 0 30 3 8 7
|
| MA0102.2_rc_shift3 (CEBPA_rc) |
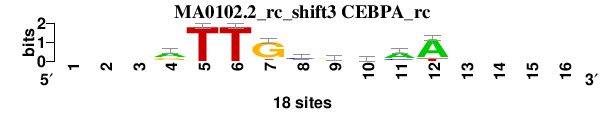 |
0.838 |
0.503 |
2.596 |
0.048 |
0.893 |
1.848 |
0.897 |
3 |
5 |
5 |
5 |
6 |
4 |
5 |
4.714 |
5 |
; oligos_7nt_mkv5_m1 versus MA0102.2_rc (CEBPA_rc); m=5/6; ncol2=9; w=9; offset=2; strand=R; shift=3; score= 4.7143; ---rTTGcrmaA----
; cor=0.838; Ncor=0.503; logoDP=2.596; NIcor=0.048; NsEucl=0.893; SSD=1.848; NSW=0.897; rcor=3; rNcor=5; rlogoDP=5; rNIcor=5; rNsEucl=6; rSSD=4; rNSW=5; rank_mean=4.714; match_rank=5
a 0 0 0 10 0 0 1 2 5 7 12 15 0 0 0 0
c 0 0 0 2 0 0 0 10 2 6 4 0 0 0 0 0
g 0 0 0 6 0 0 15 2 9 1 1 0 0 0 0 0
t 0 0 0 0 18 18 2 4 2 4 1 3 0 0 0 0
|
| MA0019.1_rc_shift0 (Ddit3::Cebpa_rc) |
 |
0.756 |
0.520 |
0.428 |
-0.161 |
0.896 |
2.597 |
0.882 |
6 |
4 |
6 |
6 |
5 |
6 |
6 |
5.571 |
6 |
; oligos_7nt_mkv5_m1 versus MA0019.1_rc (Ddit3::Cebpa_rc); m=6/6; ncol2=12; w=11; offset=-1; strand=R; shift=0; score= 5.5714; ggkATTGCAyyy----
; cor=0.756; Ncor=0.520; logoDP=0.428; NIcor=-0.161; NsEucl=0.896; SSD=2.597; NSW=0.882; rcor=6; rNcor=4; rlogoDP=6; rNIcor=6; rNsEucl=5; rSSD=6; rNSW=6; rank_mean=5.571; match_rank=6
a 7 3 3 33 0 0 2 1 38 3 7 6 0 0 0 0
c 6 9 5 0 1 0 0 38 0 15 14 12 0 0 0 0
g 26 23 17 6 2 1 33 0 1 3 7 7 0 0 0 0
t 0 4 14 0 36 38 4 0 0 18 11 14 0 0 0 0
|






