| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
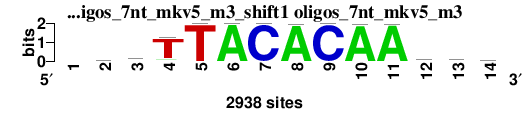
| oligos_7nt_mkv5_m3_shift1 (oligos_7nt_mkv5_m3) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_7nt_mkv5_m3; m=0 (reference); ncol1=13; shift=1; ncol=14; -wrTTACACAAtww
; Alignment reference
a 0 858 1227 211 1 2938 0 2923 0 2923 2926 729 1021 848
c 0 454 523 61 1 0 2937 0 2935 0 3 720 718 708
g 0 669 744 112 0 0 0 15 0 13 3 388 442 590
t 0 957 444 2554 2936 0 1 0 3 2 6 1101 757 792
|
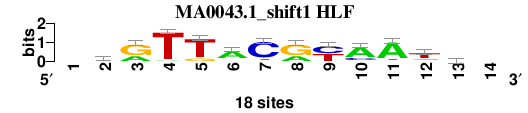
| MA0043.1_shift1 (HLF) |
 |
0.788 |
0.727 |
5.794 |
0.702 |
0.912 |
2.217 |
0.908 |
4 |
1 |
4 |
1 |
1 |
3 |
2 |
2.286 |
1 |
; oligos_7nt_mkv5_m3 versus MA0043.1 (HLF); m=1/7; ncol2=12; w=12; offset=0; strand=D; shift=1; score= 2.2857; -krTTACryAAth-
; cor=0.788; Ncor=0.727; logoDP=5.794; NIcor=0.702; NsEucl=0.912; SSD=2.217; NSW=0.908; rcor=4; rNcor=1; rlogoDP=4; rNIcor=1; rNsEucl=1; rSSD=3; rNSW=2; rank_mean=2.286; match_rank=1
a 0 1 6 1 0 13 0 6 0 13 15 2 5 0
c 0 4 0 0 0 1 15 0 9 4 0 3 5 0
g 0 8 12 0 3 2 1 12 0 1 1 1 3 0
t 0 5 0 17 15 2 2 0 9 0 2 12 5 0
|
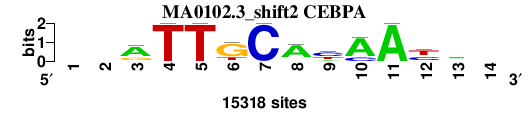
| MA0102.3_shift2 (CEBPA) |
 |
0.790 |
0.669 |
8.675 |
0.664 |
0.903 |
2.262 |
0.897 |
2 |
2 |
1 |
2 |
2 |
4 |
4 |
2.429 |
2 |
; oligos_7nt_mkv5_m3 versus MA0102.3 (CEBPA); m=2/7; ncol2=11; w=11; offset=1; strand=D; shift=2; score= 2.4286; --aTTGCAyAAym-
; cor=0.790; Ncor=0.669; logoDP=8.675; NIcor=0.664; NsEucl=0.903; SSD=2.262; NSW=0.897; rcor=2; rNcor=2; rlogoDP=1; rNIcor=2; rNsEucl=2; rSSD=4; rNSW=4; rank_mean=2.429; match_rank=2
a 0 0 10351 0 0 705 0 11956 2660 12226 15318 0 5804 0
c 0 0 1332 0 0 0 15318 1274 7654 3092 0 5688 4963 0
g 0 0 3635 0 0 11304 0 1066 0 0 0 1804 890 0
t 0 0 0 15318 15318 3309 0 1022 5004 0 0 7826 3661 0
|
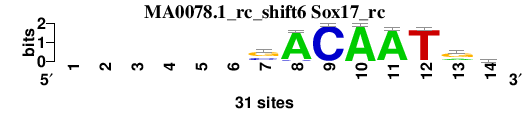
| MA0078.1_rc_shift6 (Sox17_rc) |
 |
0.799 |
0.456 |
1.990 |
0.086 |
0.890 |
1.541 |
0.904 |
1 |
6 |
7 |
6 |
5 |
2 |
3 |
4.286 |
3 |
; oligos_7nt_mkv5_m3 versus MA0078.1_rc (Sox17_rc); m=3/7; ncol2=9; w=8; offset=5; strand=R; shift=6; score= 4.2857; ------sACAATrd
; cor=0.799; Ncor=0.456; logoDP=1.990; NIcor=0.086; NsEucl=0.890; SSD=1.541; NSW=0.904; rcor=1; rNcor=6; rlogoDP=7; rNIcor=6; rNsEucl=5; rSSD=2; rNSW=3; rank_mean=4.286; match_rank=3
a 0 0 0 0 0 0 4 29 0 31 30 1 9 11
c 0 0 0 0 0 0 10 2 31 0 0 0 1 4
g 0 0 0 0 0 0 17 0 0 0 1 0 18 8
t 0 0 0 0 0 0 0 0 0 0 0 30 3 8
|
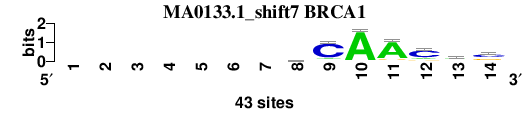
| MA0133.1_shift7 (BRCA1) |
 |
0.779 |
0.419 |
3.209 |
0.421 |
0.890 |
1.196 |
0.915 |
5 |
7 |
5 |
5 |
6 |
1 |
1 |
4.286 |
4 |
; oligos_7nt_mkv5_m3 versus MA0133.1 (BRCA1); m=4/7; ncol2=7; w=7; offset=6; strand=D; shift=7; score= 4.2857; -------mCAAcms
; cor=0.779; Ncor=0.419; logoDP=3.209; NIcor=0.421; NsEucl=0.890; SSD=1.196; NSW=0.915; rcor=5; rNcor=7; rlogoDP=5; rNIcor=5; rNsEucl=6; rSSD=1; rNSW=1; rank_mean=4.286; match_rank=4
a 0 0 0 0 0 0 0 15 4 41 36 7 19 3
c 0 0 0 0 0 0 0 11 35 1 2 29 14 22
g 0 0 0 0 0 0 0 10 2 1 4 6 7 15
t 0 0 0 0 0 0 0 7 2 0 1 1 3 3
|
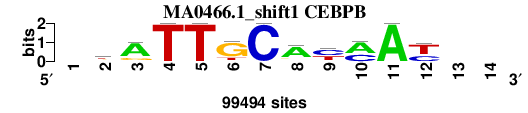
| MA0466.1_shift1 (CEBPB) |
 |
0.752 |
0.636 |
8.314 |
0.623 |
0.895 |
2.680 |
0.878 |
7 |
3 |
2 |
3 |
3 |
7 |
6 |
4.429 |
5 |
; oligos_7nt_mkv5_m3 versus MA0466.1 (CEBPB); m=5/7; ncol2=11; w=11; offset=0; strand=D; shift=1; score= 4.4286; -kATTGCAymAy--
; cor=0.752; Ncor=0.636; logoDP=8.314; NIcor=0.623; NsEucl=0.895; SSD=2.680; NSW=0.878; rcor=7; rNcor=3; rlogoDP=2; rNIcor=3; rNsEucl=3; rSSD=7; rNSW=6; rank_mean=4.429; match_rank=5
a 0 13006 75198 0 0 4556 0 74715 8654 60151 99494 0 0 0
c 0 10026 5868 0 0 0 99494 5478 51954 39343 0 36038 0 0
g 0 33617 18428 0 0 75531 0 10015 0 0 0 2043 0 0
t 0 42845 0 99494 99494 19407 0 9286 38886 0 0 61413 0 0
|
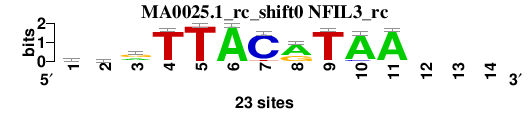
| MA0025.1_rc_shift0 (NFIL3_rc) |
 |
0.790 |
0.564 |
2.256 |
0.072 |
0.893 |
2.310 |
0.884 |
3 |
4 |
6 |
7 |
4 |
5 |
5 |
4.857 |
6 |
; oligos_7nt_mkv5_m3 versus MA0025.1_rc (NFIL3_rc); m=6/7; ncol2=11; w=10; offset=-1; strand=R; shift=0; score= 4.8571; ayrTTACrTAA---
; cor=0.790; Ncor=0.564; logoDP=2.256; NIcor=0.072; NsEucl=0.893; SSD=2.310; NSW=0.884; rcor=3; rNcor=4; rlogoDP=6; rNIcor=7; rNsEucl=4; rSSD=5; rNSW=5; rank_mean=4.857; match_rank=6
a 10 4 8 0 0 23 0 15 1 21 22 0 0 0
c 3 7 4 1 0 0 21 0 0 2 0 0 0 0
g 5 5 11 0 0 0 0 8 0 0 0 0 0 0
t 5 7 0 22 23 0 2 0 22 0 1 0 0 0
|
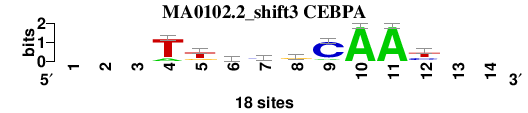
| MA0102.2_shift3 (CEBPA) |
 |
0.756 |
0.523 |
5.976 |
0.496 |
0.877 |
2.470 |
0.863 |
6 |
5 |
3 |
4 |
7 |
6 |
7 |
5.429 |
7 |
; oligos_7nt_mkv5_m3 versus MA0102.2 (CEBPA); m=7/7; ncol2=9; w=9; offset=2; strand=D; shift=3; score= 5.4286; ---TtkygCAAy--
; cor=0.756; Ncor=0.523; logoDP=5.976; NIcor=0.496; NsEucl=0.877; SSD=2.470; NSW=0.863; rcor=6; rNcor=5; rlogoDP=3; rNIcor=4; rNsEucl=7; rSSD=6; rNSW=7; rank_mean=5.429; match_rank=7
a 0 0 0 3 1 4 2 4 2 18 18 0 0 0
c 0 0 0 0 1 1 9 2 15 0 0 6 0 0
g 0 0 0 0 4 6 2 10 0 0 0 2 0 0
t 0 0 0 15 12 7 5 2 1 0 0 10 0 0
|







