| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
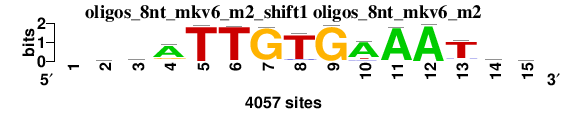
| oligos_8nt_mkv6_m2_shift1 (oligos_8nt_mkv6_m2) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_8nt_mkv6_m2; m=0 (reference); ncol1=14; shift=1; ncol=15; -wkATTGTGAAATaw
; Alignment reference
a 0 1314 898 3074 24 26 71 73 20 3257 3961 4033 93 1624 1211
c 0 660 557 241 11 10 15 263 2 360 61 7 335 985 878
g 0 993 1040 571 8 33 3952 36 4011 11 6 7 248 674 788
t 0 1090 1562 171 4014 3988 19 3685 24 429 29 10 3381 774 1180
|
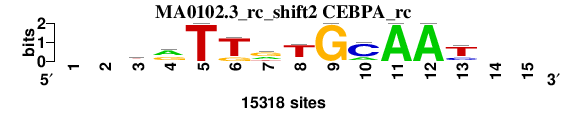
| MA0102.3_rc_shift2 (CEBPA_rc) |
 |
0.879 |
0.691 |
4.259 |
0.146 |
0.924 |
1.396 |
0.937 |
1 |
1 |
2 |
3 |
1 |
2 |
1 |
1.571 |
1 |
; oligos_8nt_mkv6_m2 versus MA0102.3_rc (CEBPA_rc); m=1/5; ncol2=11; w=11; offset=1; strand=R; shift=2; score= 1.5714; --krTTrTGCAAt--
; cor=0.879; Ncor=0.691; logoDP=4.259; NIcor=0.146; NsEucl=0.924; SSD=1.396; NSW=0.937; rcor=1; rNcor=1; rlogoDP=2; rNIcor=3; rNsEucl=1; rSSD=2; rNSW=1; rank_mean=1.571; match_rank=1
a 0 0 3661 7826 0 0 5004 1022 0 3309 15318 15318 0 0 0
c 0 0 890 1804 0 0 0 1066 0 11304 0 0 3635 0 0
g 0 0 4963 5688 0 3092 7654 1274 15318 0 0 0 1332 0 0
t 0 0 5804 0 15318 12226 2660 11956 0 705 0 0 10351 0 0
|
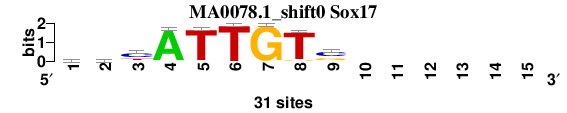
| MA0078.1_shift0 (Sox17) |
 |
0.852 |
0.454 |
6.713 |
0.449 |
0.905 |
1.143 |
0.929 |
3 |
5 |
1 |
1 |
4 |
1 |
2 |
2.429 |
2 |
; oligos_8nt_mkv6_m2 versus MA0078.1 (Sox17); m=2/5; ncol2=9; w=8; offset=-1; strand=D; shift=0; score= 2.4286; yhyATTGTs------
; cor=0.852; Ncor=0.454; logoDP=6.713; NIcor=0.449; NsEucl=0.905; SSD=1.143; NSW=0.929; rcor=3; rNcor=5; rlogoDP=1; rNIcor=1; rNsEucl=4; rSSD=1; rNSW=2; rank_mean=2.429; match_rank=2
a 7 8 3 30 0 0 0 0 0 0 0 0 0 0 0
c 9 8 18 0 1 0 0 0 17 0 0 0 0 0 0
g 6 4 1 0 0 0 31 2 10 0 0 0 0 0 0
t 9 11 9 1 30 31 0 29 4 0 0 0 0 0 0
|
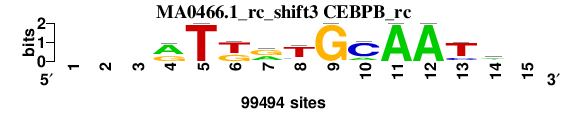
| MA0466.1_rc_shift3 (CEBPB_rc) |
 |
0.860 |
0.676 |
2.835 |
0.043 |
0.918 |
1.608 |
0.927 |
2 |
2 |
4 |
4 |
2 |
3 |
3 |
2.857 |
3 |
; oligos_8nt_mkv6_m2 versus MA0466.1_rc (CEBPB_rc); m=3/5; ncol2=11; w=11; offset=2; strand=R; shift=3; score= 2.8571; ---rTkrTGCAATm-
; cor=0.860; Ncor=0.676; logoDP=2.835; NIcor=0.043; NsEucl=0.918; SSD=1.608; NSW=0.927; rcor=2; rNcor=2; rlogoDP=4; rNIcor=4; rNsEucl=2; rSSD=3; rNSW=3; rank_mean=2.857; match_rank=3
a 0 0 0 61413 0 0 38886 9286 0 19407 99494 99494 0 42845 0
c 0 0 0 2043 0 0 0 10015 0 75531 0 0 18428 33617 0
g 0 0 0 36038 0 39343 51954 5478 99494 0 0 0 5868 10026 0
t 0 0 0 0 99494 60151 8654 74715 0 4556 0 0 75198 13006 0
|
| MA0102.2_rc_shift3 (CEBPA_rc) |
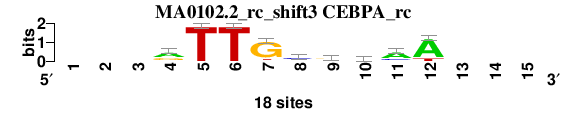 |
0.850 |
0.547 |
3.151 |
0.175 |
0.900 |
1.627 |
0.910 |
4 |
4 |
3 |
2 |
5 |
4 |
5 |
3.857 |
4 |
; oligos_8nt_mkv6_m2 versus MA0102.2_rc (CEBPA_rc); m=4/5; ncol2=9; w=9; offset=2; strand=R; shift=3; score= 3.8571; ---rTTGcrmaA---
; cor=0.850; Ncor=0.547; logoDP=3.151; NIcor=0.175; NsEucl=0.900; SSD=1.627; NSW=0.910; rcor=4; rNcor=4; rlogoDP=3; rNIcor=2; rNsEucl=5; rSSD=4; rNSW=5; rank_mean=3.857; match_rank=4
a 0 0 0 10 0 0 1 2 5 7 12 15 0 0 0
c 0 0 0 2 0 0 0 10 2 6 4 0 0 0 0
g 0 0 0 6 0 0 15 2 9 1 1 0 0 0 0
t 0 0 0 0 18 18 2 4 2 4 1 3 0 0 0
|
| MA0102.1_rc_shift0 (Cebpa_rc) |
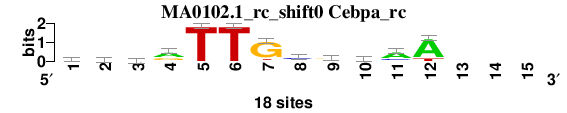 |
0.840 |
0.616 |
0.283 |
-0.189 |
0.916 |
1.717 |
0.922 |
5 |
3 |
5 |
5 |
3 |
5 |
4 |
4.286 |
5 |
; oligos_8nt_mkv6_m2 versus MA0102.1_rc (Cebpa_rc); m=5/5; ncol2=12; w=11; offset=-1; strand=R; shift=0; score= 4.2857; vrbrTTGcrmaA---
; cor=0.840; Ncor=0.616; logoDP=0.283; NIcor=-0.189; NsEucl=0.916; SSD=1.717; NSW=0.922; rcor=5; rNcor=3; rlogoDP=5; rNIcor=5; rNsEucl=3; rSSD=5; rNSW=4; rank_mean=4.286; match_rank=5
a 6 8 2 10 0 0 1 2 5 7 12 15 0 0 0
c 5 3 5 2 0 0 0 10 2 6 4 0 0 0 0
g 6 5 6 6 0 0 15 2 9 1 1 0 0 0 0
t 1 2 5 0 18 18 2 4 2 4 1 3 0 0 0
|





