One-to-n alignments
Command: compare-matrices -v 1 -mode matches -format1 transfac -file1 $RSAT/public_html/tmp/wwwrun/2014/09/03/peak-motifs.2014-09-03.094519_2014-09-03.094519_gM9GBj/results/discovered_motifs/oligos_8nt_mkv6_m3/peak-motifs_oligos_8nt_mkv6_m3.tf -format2 tf -file2 $RSAT/public_html/data/motif_databases/JASPAR/jaspar_core_vertebrates_2013-11.tf -mode matches -DR -uth offset_rank 1 -lth w 5 -lth Wr 0.3 -lth cor 0.75 -lth Ncor 0.4 -return matrix_name,matrix_id,cor,Ncor,logoDP,NIcor,NsEucl,SSD,NSW,match_rank,width,strand,offset,consensus,alignments_1ton -sort Ncor -o $RSAT/public_html/tmp/wwwrun/2014/09/03/peak-motifs.2014-09-03.094519_2014-09-03.094519_gM9GBj/results/discovered_motifs/oligos_8nt_mkv6_m3/peak-motifs_oligos_8nt_mkv6_m3_vs_db_jaspar_core_vertebrates
One-to-n matrix alignment; reference matrix: oligos_8nt_mkv6_m3_shift0 ; 5 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
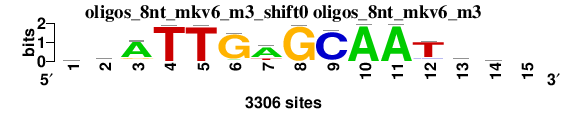
| oligos_8nt_mkv6_m3_shift0 (oligos_8nt_mkv6_m3) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_8nt_mkv6_m3; m=0 (reference); ncol1=14; shift=0; ncol=15; wtATTGAGCAATaw-
; Alignment reference
a 1047 667 2712 14 18 176 2365 4 73 3285 3285 91 1402 945 0
c 570 504 181 5 5 8 210 9 3122 9 9 294 644 732 0
g 771 667 330 10 17 3025 85 3279 5 4 4 215 531 674 0
t 918 1468 83 3277 3266 97 646 14 106 8 8 2706 729 955 0
|
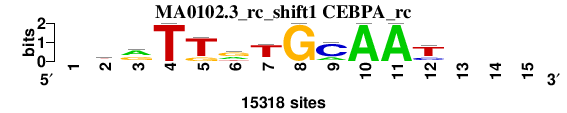
| MA0102.3_rc_shift1 (CEBPA_rc) |
 |
0.875 |
0.687 |
4.343 |
0.142 |
0.923 |
1.437 |
0.935 |
1 |
1 |
3 |
3 |
1 |
1 |
1 |
1.571 |
1 |
; oligos_8nt_mkv6_m3 versus MA0102.3_rc (CEBPA_rc); m=1/4; ncol2=11; w=11; offset=1; strand=R; shift=1; score= 1.5714; -krTTrTGCAAt---
; cor=0.875; Ncor=0.687; logoDP=4.343; NIcor=0.142; NsEucl=0.923; SSD=1.437; NSW=0.935; rcor=1; rNcor=1; rlogoDP=3; rNIcor=3; rNsEucl=1; rSSD=1; rNSW=1; rank_mean=1.571; match_rank=1
a 0 3661 7826 0 0 5004 1022 0 3309 15318 15318 0 0 0 0
c 0 890 1804 0 0 0 1066 0 11304 0 0 3635 0 0 0
g 0 4963 5688 0 3092 7654 1274 15318 0 0 0 1332 0 0 0
t 0 5804 0 15318 12226 2660 11956 0 705 0 0 10351 0 0 0
|
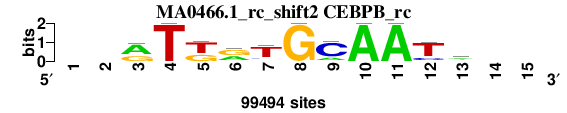
| MA0466.1_rc_shift2 (CEBPB_rc) |
 |
0.870 |
0.683 |
2.614 |
-0.034 |
0.921 |
1.491 |
0.932 |
2 |
2 |
4 |
4 |
2 |
2 |
2 |
2.571 |
2 |
; oligos_8nt_mkv6_m3 versus MA0466.1_rc (CEBPB_rc); m=2/4; ncol2=11; w=11; offset=2; strand=R; shift=2; score= 2.5714; --rTkrTGCAATm--
; cor=0.870; Ncor=0.683; logoDP=2.614; NIcor=-0.034; NsEucl=0.921; SSD=1.491; NSW=0.932; rcor=2; rNcor=2; rlogoDP=4; rNIcor=4; rNsEucl=2; rSSD=2; rNSW=2; rank_mean=2.571; match_rank=2
a 0 0 61413 0 0 38886 9286 0 19407 99494 99494 0 42845 0 0
c 0 0 2043 0 0 0 10015 0 75531 0 0 18428 33617 0 0
g 0 0 36038 0 39343 51954 5478 99494 0 0 0 5868 10026 0 0
t 0 0 0 99494 60151 8654 74715 0 4556 0 0 75198 13006 0 0
|
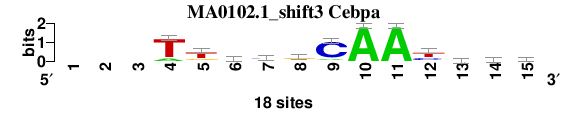
| MA0102.1_shift3 (Cebpa) |
 |
0.828 |
0.607 |
6.269 |
0.604 |
0.913 |
1.816 |
0.917 |
4 |
3 |
1 |
1 |
3 |
4 |
3 |
2.714 |
3 |
; oligos_8nt_mkv6_m3 versus MA0102.1 (Cebpa); m=3/4; ncol2=12; w=11; offset=3; strand=D; shift=3; score= 2.7143; ---TtkygCAAyvyb
; cor=0.828; Ncor=0.607; logoDP=6.269; NIcor=0.604; NsEucl=0.913; SSD=1.816; NSW=0.917; rcor=4; rNcor=3; rlogoDP=1; rNIcor=1; rNsEucl=3; rSSD=4; rNSW=3; rank_mean=2.714; match_rank=3
a 0 0 0 3 1 4 2 4 2 18 18 0 5 2 1
c 0 0 0 0 1 1 9 2 15 0 0 6 6 5 6
g 0 0 0 0 4 6 2 10 0 0 0 2 5 3 5
t 0 0 0 15 12 7 5 2 1 0 0 10 2 8 6
|
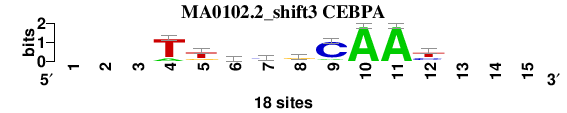
| MA0102.2_shift3 (CEBPA) |
 |
0.842 |
0.541 |
6.267 |
0.532 |
0.898 |
1.691 |
0.906 |
3 |
4 |
2 |
2 |
4 |
3 |
4 |
3.143 |
4 |
; oligos_8nt_mkv6_m3 versus MA0102.2 (CEBPA); m=4/4; ncol2=9; w=9; offset=3; strand=D; shift=3; score= 3.1429; ---TtkygCAAy---
; cor=0.842; Ncor=0.541; logoDP=6.267; NIcor=0.532; NsEucl=0.898; SSD=1.691; NSW=0.906; rcor=3; rNcor=4; rlogoDP=2; rNIcor=2; rNsEucl=4; rSSD=3; rNSW=4; rank_mean=3.143; match_rank=4
a 0 0 0 3 1 4 2 4 2 18 18 0 0 0 0
c 0 0 0 0 1 1 9 2 15 0 0 6 0 0 0
g 0 0 0 0 4 6 2 10 0 0 0 2 0 0 0
t 0 0 0 15 12 7 5 2 1 0 0 10 0 0 0
|




