| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| positions_6nt_m1_shift0 (positions_6nt_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; positions_6nt_m1; m=0 (reference); ncol1=15; shift=0; ncol=15; wwAGATTGTGAAAtm
; Alignment reference
a 358 258 771 43 867 5 8 37 26 3 721 851 886 58 368
c 124 176 20 25 8 5 2 2 40 1 82 31 1 184 243
g 173 214 31 694 17 3 17 852 7 889 6 5 6 111 102
t 242 249 75 135 5 884 870 6 824 4 88 10 4 544 184
|
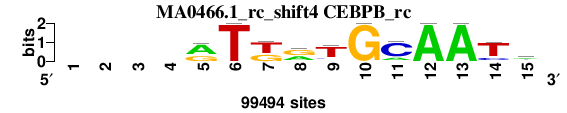
| MA0466.1_rc_shift4 (CEBPB_rc) |
 |
0.848 |
0.622 |
2.740 |
0.033 |
0.915 |
1.730 |
0.921 |
1 |
2 |
4 |
4 |
1 |
1 |
1 |
2.000 |
1 |
; positions_6nt_m1 versus MA0466.1_rc (CEBPB_rc); m=1/5; ncol2=11; w=11; offset=4; strand=R; shift=4; score= 2; ----rTkrTGCAATm
; cor=0.848; Ncor=0.622; logoDP=2.740; NIcor=0.033; NsEucl=0.915; SSD=1.730; NSW=0.921; rcor=1; rNcor=2; rlogoDP=4; rNIcor=4; rNsEucl=1; rSSD=1; rNSW=1; rank_mean=2.000; match_rank=1
a 0 0 0 0 61413 0 0 38886 9286 0 19407 99494 99494 0 42845
c 0 0 0 0 2043 0 0 0 10015 0 75531 0 0 18428 33617
g 0 0 0 0 36038 0 39343 51954 5478 99494 0 0 0 5868 10026
t 0 0 0 0 0 99494 60151 8654 74715 0 4556 0 0 75198 13006
|
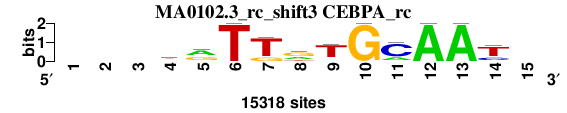
| MA0102.3_rc_shift3 (CEBPA_rc) |
 |
0.847 |
0.621 |
4.050 |
0.121 |
0.913 |
1.820 |
0.917 |
2 |
3 |
2 |
3 |
2 |
2 |
2 |
2.286 |
2 |
; positions_6nt_m1 versus MA0102.3_rc (CEBPA_rc); m=2/5; ncol2=11; w=11; offset=3; strand=R; shift=3; score= 2.2857; ---krTTrTGCAAt-
; cor=0.847; Ncor=0.621; logoDP=4.050; NIcor=0.121; NsEucl=0.913; SSD=1.820; NSW=0.917; rcor=2; rNcor=3; rlogoDP=2; rNIcor=3; rNsEucl=2; rSSD=2; rNSW=2; rank_mean=2.286; match_rank=2
a 0 0 0 3661 7826 0 0 5004 1022 0 3309 15318 15318 0 0
c 0 0 0 890 1804 0 0 0 1066 0 11304 0 0 3635 0
g 0 0 0 4963 5688 0 3092 7654 1274 15318 0 0 0 1332 0
t 0 0 0 5804 0 15318 12226 2660 11956 0 705 0 0 10351 0
|
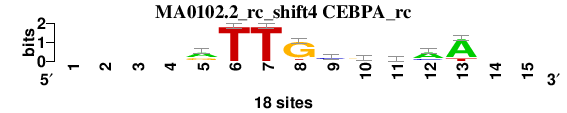
| MA0102.2_rc_shift4 (CEBPA_rc) |
 |
0.836 |
0.502 |
3.028 |
0.151 |
0.894 |
1.826 |
0.899 |
3 |
4 |
3 |
2 |
4 |
3 |
4 |
3.286 |
3 |
; positions_6nt_m1 versus MA0102.2_rc (CEBPA_rc); m=3/5; ncol2=9; w=9; offset=4; strand=R; shift=4; score= 3.2857; ----rTTGcrmaA--
; cor=0.836; Ncor=0.502; logoDP=3.028; NIcor=0.151; NsEucl=0.894; SSD=1.826; NSW=0.899; rcor=3; rNcor=4; rlogoDP=3; rNIcor=2; rNsEucl=4; rSSD=3; rNSW=4; rank_mean=3.286; match_rank=3
a 0 0 0 0 10 0 0 1 2 5 7 12 15 0 0
c 0 0 0 0 2 0 0 0 10 2 6 4 0 0 0
g 0 0 0 0 6 0 0 15 2 9 1 1 0 0 0
t 0 0 0 0 0 18 18 2 4 2 4 1 3 0 0
|
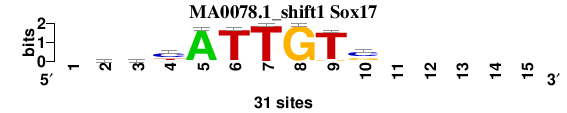
| MA0078.1_shift1 (Sox17) |
 |
0.761 |
0.456 |
7.347 |
0.462 |
0.884 |
2.168 |
0.880 |
5 |
5 |
1 |
1 |
5 |
4 |
5 |
3.714 |
4 |
; positions_6nt_m1 versus MA0078.1 (Sox17); m=4/5; ncol2=9; w=9; offset=1; strand=D; shift=1; score= 3.7143; -yhyATTGTs-----
; cor=0.761; Ncor=0.456; logoDP=7.347; NIcor=0.462; NsEucl=0.884; SSD=2.168; NSW=0.880; rcor=5; rNcor=5; rlogoDP=1; rNIcor=1; rNsEucl=5; rSSD=4; rNSW=5; rank_mean=3.714; match_rank=4
a 0 7 8 3 30 0 0 0 0 0 0 0 0 0 0
c 0 9 8 18 0 1 0 0 0 17 0 0 0 0 0
g 0 6 4 1 0 0 0 31 2 10 0 0 0 0 0
t 0 9 11 9 1 30 31 0 29 4 0 0 0 0 0
|
| MA0102.1_rc_shift1 (Cebpa_rc) |
 |
0.808 |
0.646 |
0.416 |
-0.221 |
0.908 |
2.422 |
0.899 |
4 |
1 |
5 |
5 |
3 |
5 |
3 |
3.714 |
5 |
; positions_6nt_m1 versus MA0102.1_rc (Cebpa_rc); m=5/5; ncol2=12; w=12; offset=1; strand=R; shift=1; score= 3.7143; -vrbrTTGcrmaA--
; cor=0.808; Ncor=0.646; logoDP=0.416; NIcor=-0.221; NsEucl=0.908; SSD=2.422; NSW=0.899; rcor=4; rNcor=1; rlogoDP=5; rNIcor=5; rNsEucl=3; rSSD=5; rNSW=3; rank_mean=3.714; match_rank=5
a 0 6 8 2 10 0 0 1 2 5 7 12 15 0 0
c 0 5 3 5 2 0 0 0 10 2 6 4 0 0 0
g 0 6 5 6 6 0 0 15 2 9 1 1 0 0 0
t 0 1 2 5 0 18 18 2 4 2 4 1 3 0 0
|





