| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| oligos_7nt_mkv5_m3_shift1 (oligos_7nt_mkv5_m3) |
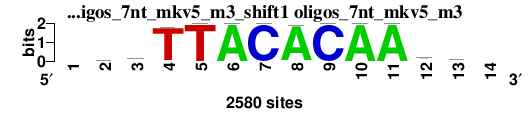 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_7nt_mkv5_m3; m=0 (reference); ncol1=12; shift=1; ncol=14; -wrTTACACAAya-
; Alignment reference
a 0 701 1073 23 1 2580 0 2542 0 2578 2579 307 920 0
c 0 409 457 6 0 0 2580 0 2580 0 1 755 634 0
g 0 616 697 30 0 0 0 38 0 2 0 388 415 0
t 0 854 353 2521 2579 0 0 0 0 0 0 1130 611 0
|
| MA0043.1_shift1 (HLF) |
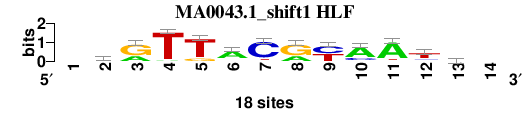 |
0.806 |
0.806 |
6.362 |
0.783 |
0.914 |
2.125 |
0.911 |
3 |
1 |
5 |
1 |
1 |
3 |
2 |
2.286 |
1 |
; oligos_7nt_mkv5_m3 versus MA0043.1 (HLF); m=1/8; ncol2=12; w=12; offset=0; strand=D; shift=1; score= 2.2857; -krTTACryAAth-
; cor=0.806; Ncor=0.806; logoDP=6.362; NIcor=0.783; NsEucl=0.914; SSD=2.125; NSW=0.911; rcor=3; rNcor=1; rlogoDP=5; rNIcor=1; rNsEucl=1; rSSD=3; rNSW=2; rank_mean=2.286; match_rank=1
a 0 1 6 1 0 13 0 6 0 13 15 2 5 0
c 0 4 0 0 0 1 15 0 9 4 0 3 5 0
g 0 8 12 0 3 2 1 12 0 1 1 1 3 0
t 0 5 0 17 15 2 2 0 9 0 2 12 5 0
|
| MA0102.3_shift2 (CEBPA) |
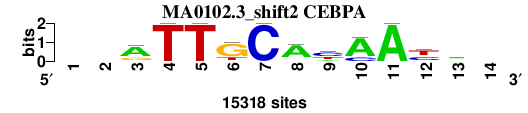 |
0.806 |
0.738 |
9.392 |
0.733 |
0.905 |
2.166 |
0.902 |
4 |
2 |
1 |
2 |
2 |
4 |
3 |
2.571 |
2 |
; oligos_7nt_mkv5_m3 versus MA0102.3 (CEBPA); m=2/8; ncol2=11; w=11; offset=1; strand=D; shift=2; score= 2.5714; --aTTGCAyAAym-
; cor=0.806; Ncor=0.738; logoDP=9.392; NIcor=0.733; NsEucl=0.905; SSD=2.166; NSW=0.902; rcor=4; rNcor=2; rlogoDP=1; rNIcor=2; rNsEucl=2; rSSD=4; rNSW=3; rank_mean=2.571; match_rank=2
a 0 0 10351 0 0 705 0 11956 2660 12226 15318 0 5804 0
c 0 0 1332 0 0 0 15318 1274 7654 3092 0 5688 4963 0
g 0 0 3635 0 0 11304 0 1066 0 0 0 1804 890 0
t 0 0 0 15318 15318 3309 0 1022 5004 0 0 7826 3661 0
|
| MA0442.1_rc_shift7 (SOX10_rc) |
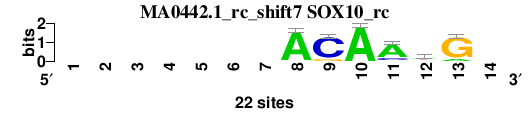 |
0.842 |
0.421 |
0.061 |
-0.129 |
0.885 |
0.948 |
0.921 |
1 |
7 |
8 |
8 |
6 |
1 |
1 |
4.571 |
3 |
; oligos_7nt_mkv5_m3 versus MA0442.1_rc (SOX10_rc); m=3/8; ncol2=6; w=6; offset=6; strand=R; shift=7; score= 4.5714; -------ACAAwG-
; cor=0.842; Ncor=0.421; logoDP=0.061; NIcor=-0.129; NsEucl=0.885; SSD=0.948; NSW=0.921; rcor=1; rNcor=7; rlogoDP=8; rNIcor=8; rNsEucl=6; rSSD=1; rNSW=1; rank_mean=4.571; match_rank=3
a 0 0 0 0 0 0 0 21 0 22 17 10 3 0
c 0 0 0 0 0 0 0 1 19 0 4 2 0 0
g 0 0 0 0 0 0 0 0 3 0 1 2 19 0
t 0 0 0 0 0 0 0 0 0 0 0 8 0 0
|
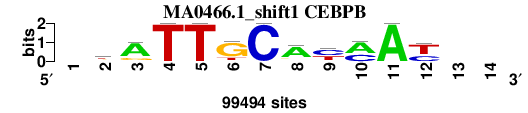
| MA0466.1_shift1 (CEBPB) |
 |
0.769 |
0.705 |
9.034 |
0.691 |
0.897 |
2.567 |
0.883 |
8 |
3 |
2 |
3 |
3 |
8 |
6 |
4.714 |
4 |
; oligos_7nt_mkv5_m3 versus MA0466.1 (CEBPB); m=4/8; ncol2=11; w=11; offset=0; strand=D; shift=1; score= 4.7143; -kATTGCAymAy--
; cor=0.769; Ncor=0.705; logoDP=9.034; NIcor=0.691; NsEucl=0.897; SSD=2.567; NSW=0.883; rcor=8; rNcor=3; rlogoDP=2; rNIcor=3; rNsEucl=3; rSSD=8; rNSW=6; rank_mean=4.714; match_rank=4
a 0 13006 75198 0 0 4556 0 74715 8654 60151 99494 0 0 0
c 0 10026 5868 0 0 0 99494 5478 51954 39343 0 36038 0 0
g 0 33617 18428 0 0 75531 0 10015 0 0 0 2043 0 0
t 0 42845 0 99494 99494 19407 0 9286 38886 0 0 61413 0 0
|
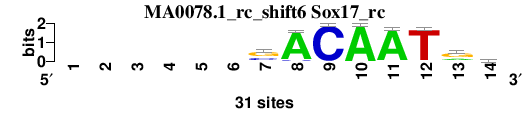
| MA0078.1_rc_shift6 (Sox17_rc) |
 |
0.814 |
0.407 |
2.071 |
0.097 |
0.879 |
1.427 |
0.898 |
2 |
8 |
7 |
6 |
7 |
2 |
4 |
5.143 |
5 |
; oligos_7nt_mkv5_m3 versus MA0078.1_rc (Sox17_rc); m=5/8; ncol2=9; w=7; offset=5; strand=R; shift=6; score= 5.1429; ------sACAATrd
; cor=0.814; Ncor=0.407; logoDP=2.071; NIcor=0.097; NsEucl=0.879; SSD=1.427; NSW=0.898; rcor=2; rNcor=8; rlogoDP=7; rNIcor=6; rNsEucl=7; rSSD=2; rNSW=4; rank_mean=5.143; match_rank=5
a 0 0 0 0 0 0 4 29 0 31 30 1 9 11
c 0 0 0 0 0 0 10 2 31 0 0 0 1 4
g 0 0 0 0 0 0 17 0 0 0 1 0 18 8
t 0 0 0 0 0 0 0 0 0 0 0 30 3 8
|
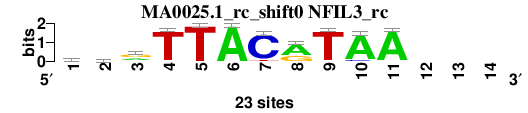
| MA0025.1_rc_shift0 (NFIL3_rc) |
 |
0.798 |
0.614 |
2.419 |
0.080 |
0.893 |
2.274 |
0.886 |
5 |
4 |
6 |
7 |
4 |
5 |
5 |
5.143 |
6 |
; oligos_7nt_mkv5_m3 versus MA0025.1_rc (NFIL3_rc); m=6/8; ncol2=11; w=10; offset=-1; strand=R; shift=0; score= 5.1429; ayrTTACrTAA---
; cor=0.798; Ncor=0.614; logoDP=2.419; NIcor=0.080; NsEucl=0.893; SSD=2.274; NSW=0.886; rcor=5; rNcor=4; rlogoDP=6; rNIcor=7; rNsEucl=4; rSSD=5; rNSW=5; rank_mean=5.143; match_rank=6
a 10 4 8 0 0 23 0 15 1 21 22 0 0 0
c 3 7 4 1 0 0 21 0 0 2 0 0 0 0
g 5 5 11 0 0 0 0 8 0 0 0 0 0 0
t 5 7 0 22 23 0 2 0 22 0 1 0 0 0
|
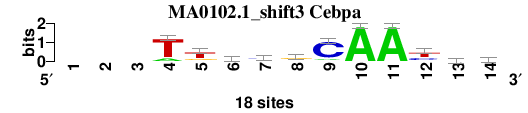
| MA0102.1_shift3 (Cebpa) |
 |
0.770 |
0.550 |
6.445 |
0.526 |
0.889 |
2.451 |
0.877 |
7 |
6 |
3 |
5 |
5 |
7 |
7 |
5.714 |
7 |
; oligos_7nt_mkv5_m3 versus MA0102.1 (Cebpa); m=7/8; ncol2=12; w=10; offset=2; strand=D; shift=3; score= 5.7143; ---TtkygCAAyvy
; cor=0.770; Ncor=0.550; logoDP=6.445; NIcor=0.526; NsEucl=0.889; SSD=2.451; NSW=0.877; rcor=7; rNcor=6; rlogoDP=3; rNIcor=5; rNsEucl=5; rSSD=7; rNSW=7; rank_mean=5.714; match_rank=7
a 0 0 0 3 1 4 2 4 2 18 18 0 5 2
c 0 0 0 0 1 1 9 2 15 0 0 6 6 5
g 0 0 0 0 4 6 2 10 0 0 0 2 5 3
t 0 0 0 15 12 7 5 2 1 0 0 10 2 8
|
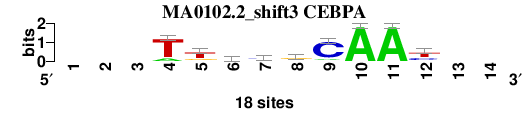
| MA0102.2_shift3 (CEBPA) |
 |
0.775 |
0.581 |
6.445 |
0.550 |
0.878 |
2.408 |
0.866 |
6 |
5 |
4 |
4 |
8 |
6 |
8 |
5.857 |
8 |
; oligos_7nt_mkv5_m3 versus MA0102.2 (CEBPA); m=8/8; ncol2=9; w=9; offset=2; strand=D; shift=3; score= 5.8571; ---TtkygCAAy--
; cor=0.775; Ncor=0.581; logoDP=6.445; NIcor=0.550; NsEucl=0.878; SSD=2.408; NSW=0.866; rcor=6; rNcor=5; rlogoDP=4; rNIcor=4; rNsEucl=8; rSSD=6; rNSW=8; rank_mean=5.857; match_rank=8
a 0 0 0 3 1 4 2 4 2 18 18 0 0 0
c 0 0 0 0 1 1 9 2 15 0 0 6 0 0
g 0 0 0 0 4 6 2 10 0 0 0 2 0 0
t 0 0 0 15 12 7 5 2 1 0 0 10 0 0
|








