One-to-n alignments
Command: compare-matrices -v 1 -mode matches -format1 transfac -file1 $RSAT/public_html/tmp/wwwrun/2014/09/03/peak-motifs.2014-09-03.182940_2014-09-03.182940_LkVj4M/results/discovered_motifs/positions_6nt_m3/peak-motifs_positions_6nt_m3.tf -format2 tf -file2 $RSAT/public_html/data/motif_databases/JASPAR/jaspar_core_vertebrates_2013-11.tf -mode matches -DR -uth offset_rank 1 -lth w 5 -lth Wr 0.3 -lth cor 0.75 -lth Ncor 0.4 -return matrix_name,matrix_id,cor,Ncor,logoDP,NIcor,NsEucl,SSD,NSW,match_rank,width,strand,offset,consensus,alignments_1ton -sort Ncor -o $RSAT/public_html/tmp/wwwrun/2014/09/03/peak-motifs.2014-09-03.182940_2014-09-03.182940_LkVj4M/results/discovered_motifs/positions_6nt_m3/peak-motifs_positions_6nt_m3_vs_db_jaspar_core_vertebrates
One-to-n matrix alignment; reference matrix: positions_6nt_m3_shift1 ; 5 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| positions_6nt_m3_shift1 (positions_6nt_m3) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; positions_6nt_m3; m=0 (reference); ncol1=13; shift=1; ncol=14; -rmAGTTGCAAAay
; Alignment reference
a 0 115 113 342 0 0 0 0 0 343 342 344 223 67
c 0 76 100 2 0 0 1 0 344 0 0 0 27 93
g 0 90 71 0 344 0 0 344 0 1 0 0 50 71
t 0 63 60 0 0 344 343 0 0 0 2 0 44 113
|
| MA0102.3_shift4 (CEBPA) |
 |
0.770 |
0.550 |
8.677 |
0.520 |
0.886 |
2.590 |
0.870 |
2 |
1 |
1 |
1 |
1 |
3 |
2 |
1.571 |
1 |
; positions_6nt_m3 versus MA0102.3 (CEBPA); m=1/4; ncol2=11; w=10; offset=3; strand=D; shift=4; score= 1.5714; ----aTTGCAyAAy
; cor=0.770; Ncor=0.550; logoDP=8.677; NIcor=0.520; NsEucl=0.886; SSD=2.590; NSW=0.870; rcor=2; rNcor=1; rlogoDP=1; rNIcor=1; rNsEucl=1; rSSD=3; rNSW=2; rank_mean=1.571; match_rank=1
a 0 0 0 0 10351 0 0 705 0 11956 2660 12226 15318 0
c 0 0 0 0 1332 0 0 0 15318 1274 7654 3092 0 5688
g 0 0 0 0 3635 0 0 11304 0 1066 0 0 0 1804
t 0 0 0 0 0 15318 15318 3309 0 1022 5004 0 0 7826
|
| MA0100.1_shift0 (Myb) |
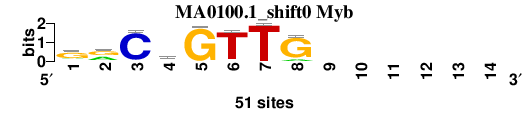 |
0.807 |
0.404 |
6.306 |
0.383 |
0.881 |
1.399 |
0.900 |
1 |
4 |
2 |
2 |
2 |
1 |
1 |
1.857 |
2 |
; positions_6nt_m3 versus MA0100.1 (Myb); m=2/4; ncol2=8; w=7; offset=-1; strand=D; shift=0; score= 1.8571; grCvGTTG------
; cor=0.807; Ncor=0.404; logoDP=6.306; NIcor=0.383; NsEucl=0.881; SSD=1.399; NSW=0.900; rcor=1; rNcor=4; rlogoDP=2; rNIcor=2; rNsEucl=2; rSSD=1; rNSW=1; rank_mean=1.857; match_rank=2
a 8 22 2 16 1 2 0 6 0 0 0 0 0 0
c 4 1 48 17 0 1 0 0 0 0 0 0 0 0
g 34 25 1 16 50 0 0 44 0 0 0 0 0 0
t 5 3 0 2 0 48 51 1 0 0 0 0 0 0
|
| MA0102.2_rc_shift4 (CEBPA_rc) |
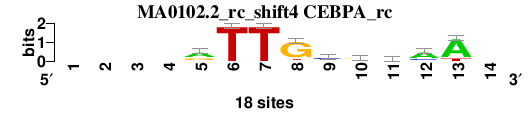 |
0.760 |
0.526 |
4.503 |
0.216 |
0.873 |
2.627 |
0.854 |
3 |
2 |
3 |
3 |
3 |
4 |
3 |
3.000 |
3 |
; positions_6nt_m3 versus MA0102.2_rc (CEBPA_rc); m=3/4; ncol2=9; w=9; offset=3; strand=R; shift=4; score= 3; ----rTTGcrmaA-
; cor=0.760; Ncor=0.526; logoDP=4.503; NIcor=0.216; NsEucl=0.873; SSD=2.627; NSW=0.854; rcor=3; rNcor=2; rlogoDP=3; rNIcor=3; rNsEucl=3; rSSD=4; rNSW=3; rank_mean=3.000; match_rank=3
a 0 0 0 0 10 0 0 1 2 5 7 12 15 0
c 0 0 0 0 2 0 0 0 10 2 6 4 0 0
g 0 0 0 0 6 0 0 15 2 9 1 1 0 0
t 0 0 0 0 0 18 18 2 4 2 4 1 3 0
|
| MA0152.1_rc_shift6 (NFATC2_rc) |
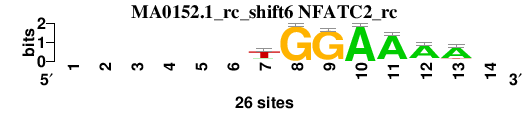 |
0.752 |
0.405 |
0.918 |
-0.050 |
0.852 |
2.141 |
0.847 |
4 |
3 |
4 |
4 |
4 |
2 |
4 |
3.571 |
4 |
; positions_6nt_m3 versus MA0152.1_rc (NFATC2_rc); m=4/4; ncol2=7; w=7; offset=5; strand=R; shift=6; score= 3.5714; ------TGGAAAA-
; cor=0.752; Ncor=0.405; logoDP=0.918; NIcor=-0.050; NsEucl=0.852; SSD=2.141; NSW=0.847; rcor=4; rNcor=3; rlogoDP=4; rNIcor=4; rNsEucl=4; rSSD=2; rNSW=4; rank_mean=3.571; match_rank=4
a 0 0 0 0 0 0 4 0 0 26 24 21 20 0
c 0 0 0 0 0 0 1 0 0 0 0 2 2 0
g 0 0 0 0 0 0 3 26 25 0 1 2 1 0
t 0 0 0 0 0 0 18 0 1 0 1 1 3 0
|




