One-to-n alignments
Command: compare-matrices -v 1 -mode matches -format1 transfac -file1 $RSAT/public_html/tmp/wwwrun/2014/09/03/peak-motifs.2014-09-03.182940_2014-09-03.182940_LkVj4M/results/discovered_motifs/positions_8nt_m2/peak-motifs_positions_8nt_m2.tf -format2 tf -file2 $RSAT/public_html/tmp/wwwrun/2014/09/03/peak-motifs.2014-09-03.182940_2014-09-03.182940_LkVj4M/peak-motifs_custom_motif_db.tf -mode matches -DR -uth offset_rank 1 -lth w 5 -lth Wr 0.3 -lth cor 0.75 -lth Ncor 0.4 -return matrix_name,matrix_id,cor,Ncor,logoDP,NIcor,NsEucl,SSD,NSW,match_rank,width,strand,offset,consensus,alignments_1ton -sort Ncor -o $RSAT/public_html/tmp/wwwrun/2014/09/03/peak-motifs.2014-09-03.182940_2014-09-03.182940_LkVj4M/results/discovered_motifs/positions_8nt_m2/peak-motifs_positions_8nt_m2_vs_db_Schmidt_CEBPa_motif
One-to-n matrix alignment; reference matrix: positions_8nt_m2_shift0 ; 2 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| positions_8nt_m2_shift0 (positions_8nt_m2) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; positions_8nt_m2; m=0 (reference); ncol1=16; shift=0; ncol=16; wrTGATTGTGCAAgad
; Alignment reference
a 183 173 43 19 589 4 1 23 18 1 24 634 632 19 271 166
c 144 131 13 20 12 4 1 4 12 1 586 4 0 72 129 147
g 140 178 24 576 24 1 10 594 7 632 3 0 4 406 107 165
t 171 156 558 23 13 629 626 17 601 4 25 0 2 141 131 160
|
| do560+do843_mmus_cebpa_liver_meme_top_m1_rc_shift3 (do560+do843_mmus_cebpa_liver_meme_top_m1_rc) |
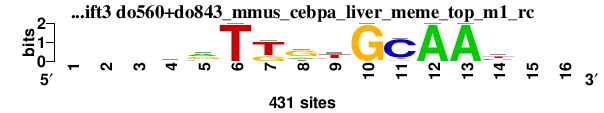 |
0.872 |
0.599 |
4.112 |
0.066 |
0.917 |
1.683 |
0.923 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1.000 |
1 |
; positions_8nt_m2 versus do560+do843_mmus_cebpa_liver_meme_top_m1_rc; m=1/1; ncol2=11; w=11; offset=3; strand=R; shift=3; score= 1; ---krTKgtGCAAy--
; cor=0.872; Ncor=0.599; logoDP=4.112; NIcor=0.066; NsEucl=0.917; SSD=1.683; NSW=0.923; rcor=1; rNcor=1; rlogoDP=1; rNIcor=1; rNsEucl=1; rSSD=1; rNSW=1; rank_mean=1.000; match_rank=1
a 0 0 0 83 183 0 1 93 39 0 31 431 431 20 0 0
c 0 0 0 60 78 0 0 0 85 0 365 0 0 138 0 0
g 0 0 0 145 169 0 130 258 48 431 0 0 0 69 0 0
t 0 0 0 143 1 431 300 80 259 0 35 0 0 204 0 0
|

