| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| positions_8nt_m2_shift1 (positions_8nt_m2) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; positions_8nt_m2; m=0 (reference); ncol1=16; shift=1; ncol=17; -wrTGATTGTGCAAgad
; Alignment reference
a 0 183 173 43 19 589 4 1 23 18 1 24 634 632 19 271 166
c 0 144 131 13 20 12 4 1 4 12 1 586 4 0 72 129 147
g 0 140 178 24 576 24 1 10 594 7 632 3 0 4 406 107 165
t 0 171 156 558 23 13 629 626 17 601 4 25 0 2 141 131 160
|
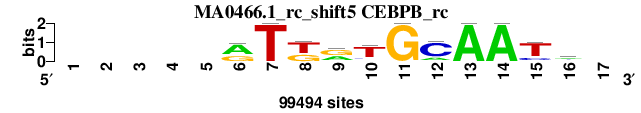
| MA0466.1_rc_shift5 (CEBPB_rc) |
 |
0.871 |
0.599 |
2.668 |
-0.037 |
0.920 |
1.536 |
0.930 |
1 |
1 |
4 |
5 |
1 |
1 |
1 |
2.000 |
1 |
; positions_8nt_m2 versus MA0466.1_rc (CEBPB_rc); m=1/6; ncol2=11; w=11; offset=4; strand=R; shift=5; score= 2; -----rTkrTGCAATm-
; cor=0.871; Ncor=0.599; logoDP=2.668; NIcor=-0.037; NsEucl=0.920; SSD=1.536; NSW=0.930; rcor=1; rNcor=1; rlogoDP=4; rNIcor=5; rNsEucl=1; rSSD=1; rNSW=1; rank_mean=2.000; match_rank=1
a 0 0 0 0 0 61413 0 0 38886 9286 0 19407 99494 99494 0 42845 0
c 0 0 0 0 0 2043 0 0 0 10015 0 75531 0 0 18428 33617 0
g 0 0 0 0 0 36038 0 39343 51954 5478 99494 0 0 0 5868 10026 0
t 0 0 0 0 0 0 99494 60151 8654 74715 0 4556 0 0 75198 13006 0
|
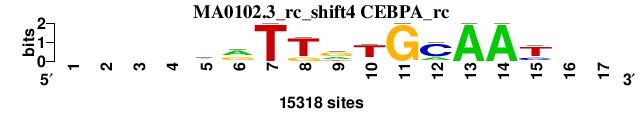
| MA0102.3_rc_shift4 (CEBPA_rc) |
 |
0.862 |
0.593 |
4.265 |
0.040 |
0.914 |
1.774 |
0.919 |
2 |
2 |
3 |
4 |
2 |
3 |
2 |
2.571 |
2 |
; positions_8nt_m2 versus MA0102.3_rc (CEBPA_rc); m=2/6; ncol2=11; w=11; offset=3; strand=R; shift=4; score= 2.5714; ----krTTrTGCAAt--
; cor=0.862; Ncor=0.593; logoDP=4.265; NIcor=0.040; NsEucl=0.914; SSD=1.774; NSW=0.919; rcor=2; rNcor=2; rlogoDP=3; rNIcor=4; rNsEucl=2; rSSD=3; rNSW=2; rank_mean=2.571; match_rank=2
a 0 0 0 0 3661 7826 0 0 5004 1022 0 3309 15318 15318 0 0 0
c 0 0 0 0 890 1804 0 0 0 1066 0 11304 0 0 3635 0 0
g 0 0 0 0 4963 5688 0 3092 7654 1274 15318 0 0 0 1332 0 0
t 0 0 0 0 5804 0 15318 12226 2660 11956 0 705 0 0 10351 0 0
|
| MA0102.1_shift6 (Cebpa) |
 |
0.780 |
0.505 |
6.062 |
0.491 |
0.904 |
2.253 |
0.898 |
4 |
3 |
2 |
1 |
3 |
5 |
4 |
3.143 |
3 |
; positions_8nt_m2 versus MA0102.1 (Cebpa); m=3/6; ncol2=12; w=11; offset=5; strand=D; shift=6; score= 3.1429; ------TtkygCAAyvy
; cor=0.780; Ncor=0.505; logoDP=6.062; NIcor=0.491; NsEucl=0.904; SSD=2.253; NSW=0.898; rcor=4; rNcor=3; rlogoDP=2; rNIcor=1; rNsEucl=3; rSSD=5; rNSW=4; rank_mean=3.143; match_rank=3
a 0 0 0 0 0 0 3 1 4 2 4 2 18 18 0 5 2
c 0 0 0 0 0 0 0 1 1 9 2 15 0 0 6 6 5
g 0 0 0 0 0 0 0 4 6 2 10 0 0 0 2 5 3
t 0 0 0 0 0 0 15 12 7 5 2 1 0 0 10 2 8
|
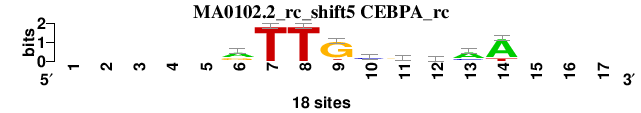
| MA0102.2_rc_shift5 (CEBPA_rc) |
 |
0.812 |
0.457 |
2.608 |
0.048 |
0.885 |
2.126 |
0.882 |
3 |
4 |
5 |
3 |
5 |
4 |
5 |
4.143 |
4 |
; positions_8nt_m2 versus MA0102.2_rc (CEBPA_rc); m=4/6; ncol2=9; w=9; offset=4; strand=R; shift=5; score= 4.1429; -----rTTGcrmaA---
; cor=0.812; Ncor=0.457; logoDP=2.608; NIcor=0.048; NsEucl=0.885; SSD=2.126; NSW=0.882; rcor=3; rNcor=4; rlogoDP=5; rNIcor=3; rNsEucl=5; rSSD=4; rNSW=5; rank_mean=4.143; match_rank=4
a 0 0 0 0 0 10 0 0 1 2 5 7 12 15 0 0 0
c 0 0 0 0 0 2 0 0 0 10 2 6 4 0 0 0 0
g 0 0 0 0 0 6 0 0 15 2 9 1 1 0 0 0 0
t 0 0 0 0 0 0 18 18 2 4 2 4 1 3 0 0 0
|
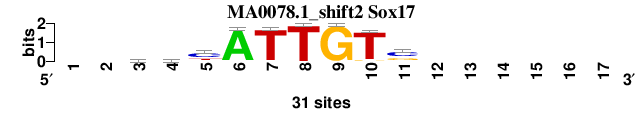
| MA0078.1_shift2 (Sox17) |
 |
0.752 |
0.423 |
7.158 |
0.424 |
0.881 |
2.296 |
0.872 |
6 |
5 |
1 |
2 |
6 |
6 |
6 |
4.571 |
5 |
; positions_8nt_m2 versus MA0078.1 (Sox17); m=5/6; ncol2=9; w=9; offset=1; strand=D; shift=2; score= 4.5714; --yhyATTGTs------
; cor=0.752; Ncor=0.423; logoDP=7.158; NIcor=0.424; NsEucl=0.881; SSD=2.296; NSW=0.872; rcor=6; rNcor=5; rlogoDP=1; rNIcor=2; rNsEucl=6; rSSD=6; rNSW=6; rank_mean=4.571; match_rank=5
a 0 0 7 8 3 30 0 0 0 0 0 0 0 0 0 0 0
c 0 0 9 8 18 0 1 0 0 0 17 0 0 0 0 0 0
g 0 0 6 4 1 0 0 0 31 2 10 0 0 0 0 0 0
t 0 0 9 11 9 1 30 31 0 29 4 0 0 0 0 0 0
|
| MA0038.1_rc_shift0 (Gfi1_rc) |
 |
0.779 |
0.413 |
0.367 |
-0.139 |
0.895 |
1.770 |
0.902 |
5 |
6 |
6 |
6 |
4 |
2 |
3 |
4.571 |
6 |
; positions_8nt_m2 versus MA0038.1_rc (Gfi1_rc); m=6/6; ncol2=10; w=9; offset=-1; strand=R; shift=0; score= 4.5714; cwgtGATTkr-------
; cor=0.779; Ncor=0.413; logoDP=0.367; NIcor=-0.139; NsEucl=0.895; SSD=1.770; NSW=0.902; rcor=5; rNcor=6; rlogoDP=6; rNIcor=6; rNsEucl=4; rSSD=2; rNSW=3; rank_mean=4.571; match_rank=6
a 5 21 6 13 0 51 0 0 2 15 0 0 0 0 0 0 0
c 28 2 11 2 0 1 0 0 7 9 0 0 0 0 0 0 0
g 13 11 28 7 52 0 0 0 16 20 0 0 0 0 0 0 0
t 7 19 8 31 1 1 53 53 28 9 0 0 0 0 0 0 0
|






