One-to-n alignments
Command: compare-matrices -v 1 -mode matches -format1 transfac -file1 $RSAT/public_html/tmp/wwwrun/2014/09/06/peak-motifs.2014-09-06.123915_2014-09-06.123915_sfIm4F/results/discovered_motifs/oligos_6nt_test_vs_ctrl_m2/peak-motifs_oligos_6nt_test_vs_ctrl_m2.tf -format2 tf -file2 $RSAT/public_html/data/motif_databases/JASPAR/jaspar_core_vertebrates_2013-11.tf -mode matches -DR -uth offset_rank 1 -lth w 5 -lth Wr 0.3 -lth cor 0.75 -lth Ncor 0.4 -return matrix_name,matrix_id,cor,Ncor,logoDP,NIcor,NsEucl,SSD,NSW,match_rank,width,strand,offset,consensus,alignments_1ton -sort Ncor -o $RSAT/public_html/tmp/wwwrun/2014/09/06/peak-motifs.2014-09-06.123915_2014-09-06.123915_sfIm4F/results/discovered_motifs/oligos_6nt_test_vs_ctrl_m2/peak-motifs_oligos_6nt_test_vs_ctrl_m2_vs_db_jaspar_core_vertebrates
One-to-n matrix alignment; reference matrix: oligos_6nt_test_vs_ctrl_m2_shift2 ; 2 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
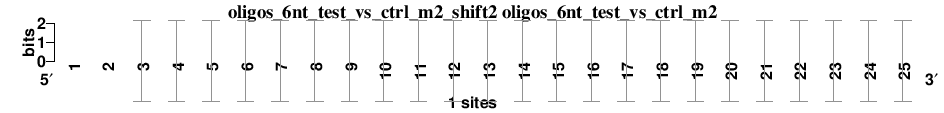
| oligos_6nt_test_vs_ctrl_m2_shift2 (oligos_6nt_test_vs_ctrl_m2) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_6nt_test_vs_ctrl_m2; m=0 (reference); ncol1=23; shift=2; ncol=25; --ATCCAAAGTCCAGAGCAGGGAAT
; Alignment reference
a 0 0 1 0 0 0 1 1 1 0 0 0 0 1 0 1 0 0 1 0 0 0 1 1 0
c 0 0 0 0 1 1 0 0 0 0 0 1 1 0 0 0 0 1 0 0 0 0 0 0 0
g 0 0 0 0 0 0 0 0 0 1 0 0 0 0 1 0 1 0 0 1 1 1 0 0 0
t 0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 1
|
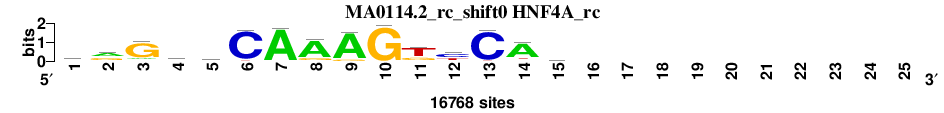
| MA0114.2_rc_shift0 (HNF4A_rc) |
 |
0.774 |
0.402 |
4.452 |
0.058 |
0.892 |
3.946 |
0.848 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1.000 |
1 |
; oligos_6nt_test_vs_ctrl_m2 versus MA0114.2_rc (HNF4A_rc); m=1/1; ncol2=15; w=13; offset=-2; strand=R; shift=0; score= 1; rrGkyCAAAGkyCAg----------
; cor=0.774; Ncor=0.402; logoDP=4.452; NIcor=0.058; NsEucl=0.892; SSD=3.946; NSW=0.848; rcor=1; rNcor=1; rlogoDP=1; rNIcor=1; rNsEucl=1; rSSD=1; rNSW=1; rank_mean=1.000; match_rank=1
a 5247 7985 1769 3334 2413 128 16188 13586 15112 49 988 477 70 13410 4057 0 0 0 0 0 0 0 0 0 0
c 3001 415 352 1662 5340 15741 407 367 0 67 329 8370 15810 907 3974 0 0 0 0 0 0 0 0 0 0
g 6450 6498 13491 5653 3745 35 173 2637 1630 16535 4425 1846 51 720 5971 0 0 0 0 0 0 0 0 0 0
t 2070 1870 1156 6119 5270 864 0 178 26 117 11026 6075 837 1731 2766 0 0 0 0 0 0 0 0 0 0
|

