One-to-n alignments
Command: compare-matrices -v 1 -mode matches -format1 transfac -file1 $RSAT/public_html/tmp/wwwrun/2014/09/06/peak-motifs.2014-09-06.123915_2014-09-06.123915_sfIm4F/results/discovered_motifs/oligos_8nt_test_vs_ctrl_m5/peak-motifs_oligos_8nt_test_vs_ctrl_m5.tf -format2 tf -file2 $RSAT/public_html/data/motif_databases/JASPAR/jaspar_core_vertebrates_2013-11.tf -mode matches -DR -uth offset_rank 1 -lth w 5 -lth Wr 0.3 -lth cor 0.75 -lth Ncor 0.4 -return matrix_name,matrix_id,cor,Ncor,logoDP,NIcor,NsEucl,SSD,NSW,match_rank,width,strand,offset,consensus,alignments_1ton -sort Ncor -o $RSAT/public_html/tmp/wwwrun/2014/09/06/peak-motifs.2014-09-06.123915_2014-09-06.123915_sfIm4F/results/discovered_motifs/oligos_8nt_test_vs_ctrl_m5/peak-motifs_oligos_8nt_test_vs_ctrl_m5_vs_db_jaspar_core_vertebrates
One-to-n matrix alignment; reference matrix: oligos_8nt_test_vs_ctrl_m5_shift4 ; 4 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| oligos_8nt_test_vs_ctrl_m5_shift4 (oligos_8nt_test_vs_ctrl_m5) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_8nt_test_vs_ctrl_m5; m=0 (reference); ncol1=13; shift=4; ncol=17; ----aaAAATCAATAww
; Alignment reference
a 0 0 0 0 185 190 438 449 466 1 13 425 465 0 467 139 167
c 0 0 0 0 92 81 4 0 0 0 452 0 1 1 2 93 76
g 0 0 0 0 94 90 8 4 2 0 0 29 1 0 0 102 81
t 0 0 0 0 98 108 19 16 1 468 4 15 2 468 0 135 145
|
| MA0594.1_shift1 (Hoxa9) |
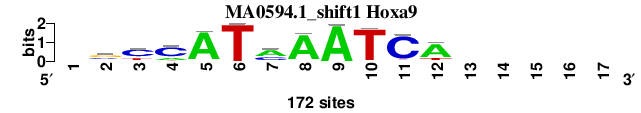 |
0.826 |
0.413 |
6.314 |
0.388 |
0.891 |
1.508 |
0.906 |
1 |
2 |
2 |
2 |
2 |
1 |
1 |
1.571 |
1 |
; oligos_8nt_test_vs_ctrl_m5 versus MA0594.1 (Hoxa9); m=1/3; ncol2=11; w=8; offset=-3; strand=D; shift=1; score= 1.5714; -syCATmAATCA-----
; cor=0.826; Ncor=0.413; logoDP=6.314; NIcor=0.388; NsEucl=0.891; SSD=1.508; NSW=0.906; rcor=1; rNcor=2; rlogoDP=2; rNIcor=2; rNsEucl=2; rSSD=1; rNSW=1; rank_mean=1.571; match_rank=1
a 0 34 12 38 161 0 101 157 171 5 7 131 0 0 0 0 0
c 0 53 110 117 3 0 58 6 1 0 154 11 0 0 0 0 0
g 0 81 5 0 8 0 8 0 0 1 0 0 0 0 0 0 0
t 0 4 45 17 0 172 5 9 0 166 11 30 0 0 0 0 0
|
| MA0047.1_rc_shift6 (Foxa2_rc) |
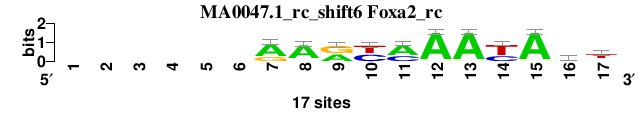 |
0.787 |
0.618 |
3.193 |
0.211 |
0.900 |
2.399 |
0.891 |
2 |
1 |
3 |
3 |
1 |
3 |
2 |
2.143 |
2 |
; oligos_8nt_test_vs_ctrl_m5 versus MA0047.1_rc (Foxa2_rc); m=2/3; ncol2=12; w=11; offset=2; strand=R; shift=6; score= 2.1429; ------RArymAAyAwt
; cor=0.787; Ncor=0.618; logoDP=3.193; NIcor=0.211; NsEucl=0.900; SSD=2.399; NSW=0.891; rcor=2; rNcor=1; rlogoDP=3; rNIcor=3; rNsEucl=1; rSSD=3; rNSW=2; rank_mean=2.143; match_rank=2
a 0 0 0 0 0 0 12 14 8 0 11 16 16 0 16 5 2
c 0 0 0 0 0 0 0 1 0 8 6 0 0 6 0 1 1
g 0 0 0 0 0 0 5 1 9 0 0 1 1 0 1 3 3
t 0 0 0 0 0 0 0 1 0 9 0 0 0 11 0 8 11
|
| MA0485.1_shift0 (Hoxc9) |
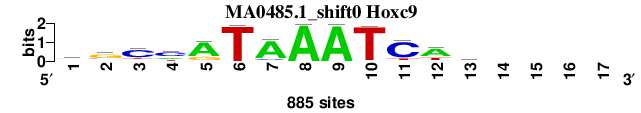 |
0.770 |
0.407 |
6.798 |
0.388 |
0.885 |
2.142 |
0.881 |
3 |
3 |
1 |
1 |
3 |
2 |
3 |
2.286 |
3 |
; oligos_8nt_test_vs_ctrl_m5 versus MA0485.1 (Hoxc9); m=3/3; ncol2=13; w=9; offset=-4; strand=D; shift=0; score= 2.2857; rgCmATAAATCwy----
; cor=0.770; Ncor=0.407; logoDP=6.798; NIcor=0.388; NsEucl=0.885; SSD=2.142; NSW=0.881; rcor=3; rNcor=3; rlogoDP=1; rNIcor=1; rNsEucl=3; rSSD=2; rNSW=3; rank_mean=2.286; match_rank=3
a 235 112 88 224 661 0 745 881 879 18 48 567 141 0 0 0 0
c 110 171 605 468 3 9 84 0 6 0 715 72 329 0 0 0 0
g 401 536 40 1 205 0 35 0 0 0 5 6 160 0 0 0 0
t 139 66 152 192 16 876 21 4 0 867 117 240 255 0 0 0 0
|



