One-to-n alignments
Command: compare-matrices -v 1 -mode matches -format1 transfac -file1 $RSAT/public_html/tmp/apache/2014/09/05/peak-motifs.2014-09-05.194903_2014-09-05.194903_oebJl9/results/discovered_motifs/positions_6nt_m1/peak-motifs_positions_6nt_m1.tf -format2 tf -file2 $RSAT/public_html/data/motif_databases/JASPAR/jaspar_core_vertebrates_2013-11.tf -mode matches -DR -uth offset_rank 1 -lth w 5 -lth Wr 0.3 -lth cor 0.75 -lth Ncor 0.4 -return matrix_name,matrix_id,cor,Ncor,logoDP,NIcor,NsEucl,SSD,NSW,match_rank,width,strand,offset,consensus,alignments_1ton -sort Ncor -o $RSAT/public_html/tmp/apache/2014/09/05/peak-motifs.2014-09-05.194903_2014-09-05.194903_oebJl9/results/discovered_motifs/positions_6nt_m1/peak-motifs_positions_6nt_m1_vs_db_jaspar_core_vertebrates
One-to-n matrix alignment; reference matrix: positions_6nt_m1_shift2 ; 5 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
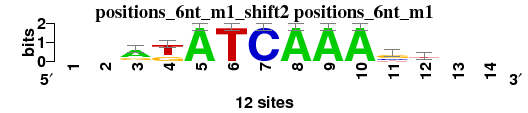
| positions_6nt_m1_shift2 (positions_6nt_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; positions_6nt_m1; m=0 (reference); ncol1=10; shift=2; ncol=14; --rkATCAAAsd--
; Alignment reference
a 0 0 8 0 12 0 0 12 12 12 0 4 0 0
c 0 0 1 0 0 0 12 0 0 0 3 0 0 0
g 0 0 3 4 0 0 0 0 0 0 7 3 0 0
t 0 0 0 8 0 12 0 0 0 0 2 5 0 0
|
| MA0523.1_shift0 (TCF7L2) |
 |
0.864 |
0.617 |
9.193 |
0.634 |
0.917 |
1.388 |
0.931 |
2 |
1 |
1 |
1 |
1 |
3 |
1 |
1.429 |
1 |
; positions_6nt_m1 versus MA0523.1 (TCF7L2); m=1/4; ncol2=14; w=10; offset=-2; strand=D; shift=0; score= 1.4286; rrAswTCAAAGgva
; cor=0.864; Ncor=0.617; logoDP=9.193; NIcor=0.634; NsEucl=0.917; SSD=1.388; NSW=0.931; rcor=2; rNcor=1; rlogoDP=1; rNIcor=1; rNsEucl=1; rSSD=3; rNSW=1; rank_mean=1.429; match_rank=1
a 1525 1808 3070 85 2795 138 155 4188 4103 4169 303 1035 1398 1782
c 756 840 136 1947 26 0 3304 0 15 0 37 849 1123 826
g 1140 1153 559 2008 0 0 729 0 70 19 3848 2008 1210 757
t 767 387 423 148 1367 4050 0 0 0 0 0 296 457 823
|
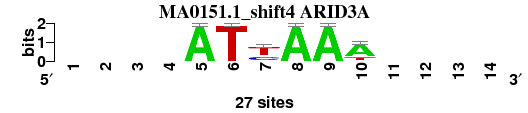
| MA0151.1_shift4 (ARID3A) |
 |
0.887 |
0.532 |
8.694 |
0.516 |
0.885 |
0.960 |
0.920 |
1 |
2 |
2 |
2 |
2 |
1 |
2 |
1.714 |
2 |
; positions_6nt_m1 versus MA0151.1 (ARID3A); m=2/4; ncol2=6; w=6; offset=2; strand=D; shift=4; score= 1.7143; ----ATyAAA----
; cor=0.887; Ncor=0.532; logoDP=8.694; NIcor=0.516; NsEucl=0.885; SSD=0.960; NSW=0.920; rcor=1; rNcor=2; rlogoDP=2; rNIcor=2; rNsEucl=2; rSSD=1; rNSW=2; rank_mean=1.714; match_rank=2
a 0 0 0 0 27 0 1 27 27 20 0 0 0 0
c 0 0 0 0 0 0 9 0 0 0 0 0 0 0
g 0 0 0 0 0 0 0 0 0 1 0 0 0 0
t 0 0 0 0 0 27 17 0 0 6 0 0 0 0
|
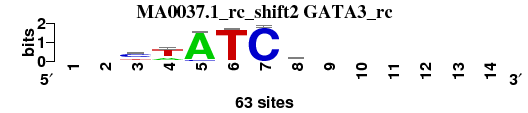
| MA0037.1_rc_shift2 (GATA3_rc) |
 |
0.759 |
0.455 |
3.521 |
0.251 |
0.854 |
1.532 |
0.872 |
4 |
3 |
3 |
3 |
3 |
4 |
4 |
3.429 |
3 |
; positions_6nt_m1 versus MA0037.1_rc (GATA3_rc); m=3/4; ncol2=6; w=6; offset=0; strand=R; shift=2; score= 3.4286; --cwATCw------
; cor=0.759; Ncor=0.455; logoDP=3.521; NIcor=0.251; NsEucl=0.854; SSD=1.532; NSW=0.872; rcor=4; rNcor=3; rlogoDP=3; rNIcor=3; rNsEucl=3; rSSD=4; rNSW=4; rank_mean=3.429; match_rank=3
a 0 0 8 19 58 1 0 20 0 0 0 0 0 0
c 0 0 37 4 5 1 62 4 0 0 0 0 0 0
g 0 0 3 1 0 0 1 14 0 0 0 0 0 0
t 0 0 15 39 0 61 0 25 0 0 0 0 0 0
|
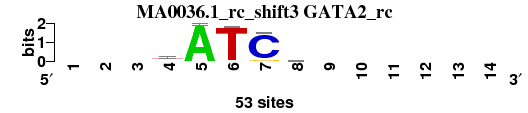
| MA0036.1_rc_shift3 (GATA2_rc) |
 |
0.814 |
0.407 |
0.110 |
-0.082 |
0.851 |
1.115 |
0.889 |
3 |
4 |
4 |
4 |
4 |
2 |
3 |
3.429 |
4 |
; positions_6nt_m1 versus MA0036.1_rc (GATA2_rc); m=4/4; ncol2=5; w=5; offset=1; strand=R; shift=3; score= 3.4286; ---yATCc------
; cor=0.814; Ncor=0.407; logoDP=0.110; NIcor=-0.082; NsEucl=0.851; SSD=1.115; NSW=0.889; rcor=3; rNcor=4; rlogoDP=4; rNIcor=4; rNsEucl=4; rSSD=2; rNSW=3; rank_mean=3.429; match_rank=4
a 0 0 0 6 53 0 0 9 0 0 0 0 0 0
c 0 0 0 15 0 1 48 18 0 0 0 0 0 0
g 0 0 0 7 0 0 5 13 0 0 0 0 0 0
t 0 0 0 25 0 52 0 13 0 0 0 0 0 0
|




