One-to-n alignments
Command: compare-matrices -v 1 -mode matches -format1 transfac -file1 $RSAT/public_html/tmp/apache/2014/09/05/peak-motifs.2014-09-05.194903_2014-09-05.194903_oebJl9/results/discovered_motifs/positions_8nt_m5/peak-motifs_positions_8nt_m5.tf -format2 tf -file2 $RSAT/public_html/data/motif_databases/JASPAR/jaspar_core_vertebrates_2013-11.tf -mode matches -DR -uth offset_rank 1 -lth w 5 -lth Wr 0.3 -lth cor 0.75 -lth Ncor 0.4 -return matrix_name,matrix_id,cor,Ncor,logoDP,NIcor,NsEucl,SSD,NSW,match_rank,width,strand,offset,consensus,alignments_1ton -sort Ncor -o $RSAT/public_html/tmp/apache/2014/09/05/peak-motifs.2014-09-05.194903_2014-09-05.194903_oebJl9/results/discovered_motifs/positions_8nt_m5/peak-motifs_positions_8nt_m5_vs_db_jaspar_core_vertebrates
One-to-n matrix alignment; reference matrix: positions_8nt_m5_shift0 ; 2 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| positions_8nt_m5_shift0 (positions_8nt_m5) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; positions_8nt_m5; m=0 (reference); ncol1=12; shift=0; ncol=12; dtAACCCGGAtg
; Alignment reference
a 13 6 33 39 0 1 1 1 7 36 9 9
c 6 7 2 2 43 42 38 0 0 2 8 8
g 12 8 3 1 0 0 0 41 31 2 10 18
t 12 22 5 1 0 0 4 1 5 3 16 8
|
| MA0098.1_rc_shift6 (ETS1_rc) |
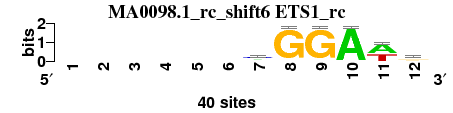 |
0.841 |
0.421 |
0.238 |
-0.094 |
0.898 |
0.750 |
0.937 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1.000 |
1 |
; positions_8nt_m5 versus MA0098.1_rc (ETS1_rc); m=1/1; ncol2=6; w=6; offset=6; strand=R; shift=6; score= 1; ------mGGAwr
; cor=0.841; Ncor=0.421; logoDP=0.238; NIcor=-0.094; NsEucl=0.898; SSD=0.750; NSW=0.937; rcor=1; rNcor=1; rlogoDP=1; rNIcor=1; rNsEucl=1; rSSD=1; rNSW=1; rank_mean=1.000; match_rank=1
a 0 0 0 0 0 0 15 1 0 39 23 16
c 0 0 0 0 0 0 17 0 1 0 0 4
g 0 0 0 0 0 0 3 39 39 1 0 16
t 0 0 0 0 0 0 5 0 0 0 17 4
|

