| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
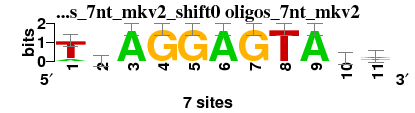
| oligos_7nt_mkv2_shift0 (oligos_7nt_mkv2) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_7nt_mkv2; m=0 (reference); ncol1=11; shift=0; ncol=11; TsAGGAGTAhw
; Alignment reference
a 1 1 7 0 0 7 0 0 7 2 3
c 0 2 0 0 0 0 0 0 0 2 0
g 0 3 0 7 7 0 7 0 0 0 1
t 6 1 0 0 0 0 0 7 0 3 3
|
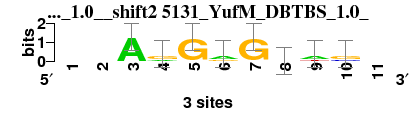
| 5131_YufM_DBTBS_1.0__shift2 (5131_YufM_DBTBS_1.0_) |
 |
0.786 |
0.572 |
7.999 |
0.589 |
0.874 |
2.045 |
0.872 |
3 |
2 |
2 |
1 |
3 |
6 |
3 |
2.857 |
1 |
; oligos_7nt_mkv2 versus 5131_YufM_DBTBS_1.0_; m=1/7; ncol2=8; w=8; offset=2; strand=D; shift=2; score= 2.8571; --ArGrGdws-
; cor=0.786; Ncor=0.572; logoDP=7.999; NIcor=0.589; NsEucl=0.874; SSD=2.045; NSW=0.872; rcor=3; rNcor=2; rlogoDP=2; rNIcor=1; rNsEucl=3; rSSD=6; rNSW=3; rank_mean=2.857; match_rank=1
a 0 0 3 1 0 2 0 1 2 0 0
c 0 0 0 0 0 0 0 0 0 1 0
g 0 0 0 2 3 1 3 1 0 2 0
t 0 0 0 0 0 0 0 1 1 0 0
|
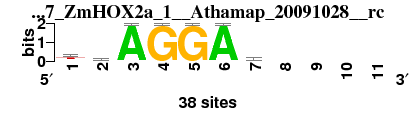
| 5047_ZmHOX2a_1__Athamap_20091028__rc_shift0 (5047_ZmHOX2a_1__Athamap_20091028__rc) |
 |
0.825 |
0.525 |
0.129 |
-0.139 |
0.881 |
1.382 |
0.901 |
1 |
4 |
7 |
6 |
1 |
1 |
1 |
3.000 |
2 |
; oligos_7nt_mkv2 versus 5047_ZmHOX2a_1__Athamap_20091028__rc; m=2/7; ncol2=7; w=7; offset=0; strand=R; shift=0; score= 3; ttAGGAy----
; cor=0.825; Ncor=0.525; logoDP=0.129; NIcor=-0.139; NsEucl=0.881; SSD=1.382; NSW=0.901; rcor=1; rNcor=4; rlogoDP=7; rNIcor=6; rNsEucl=1; rSSD=1; rNSW=1; rank_mean=3.000; match_rank=2
a 8 6 38 0 0 38 7 0 0 0 0
c 3 8 0 0 0 0 16 0 0 0 0
g 6 7 0 38 38 0 4 0 0 0 0
t 21 17 0 0 0 0 11 0 0 0 0
|
| 2780_PF0093.1_JASPAR_CORE_2009__shift3 (2780_PF0093.1_JASPAR_CORE_2009_) |
 |
0.778 |
0.424 |
9.609 |
0.424 |
0.833 |
2.000 |
0.833 |
4 |
5 |
1 |
3 |
5 |
2 |
5 |
3.571 |
3 |
; oligos_7nt_mkv2 versus 2780_PF0093.1_JASPAR_CORE_2009_; m=3/7; ncol2=6; w=6; offset=3; strand=D; shift=3; score= 3.5714; ---GGATTA--
; cor=0.778; Ncor=0.424; logoDP=9.609; NIcor=0.424; NsEucl=0.833; SSD=2.000; NSW=0.833; rcor=4; rNcor=5; rlogoDP=1; rNIcor=3; rNsEucl=5; rSSD=2; rNSW=5; rank_mean=3.571; match_rank=3
a 0 0 0 0 0 1992 0 0 1992 0 0
c 0 0 0 0 0 0 0 0 0 0 0
g 0 0 0 1992 1992 0 0 0 0 0 0
t 0 0 0 0 0 0 1992 1992 0 0 0
|
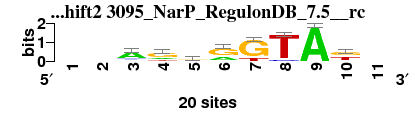
| 3095_NarP_RegulonDB_7.5__rc_shift2 (3095_NarP_RegulonDB_7.5__rc) |
 |
0.789 |
0.574 |
0.939 |
-0.161 |
0.874 |
2.044 |
0.872 |
2 |
1 |
6 |
7 |
2 |
5 |
2 |
3.571 |
4 |
; oligos_7nt_mkv2 versus 3095_NarP_RegulonDB_7.5__rc; m=4/7; ncol2=8; w=8; offset=2; strand=R; shift=2; score= 3.5714; --AgrrGTAk-
; cor=0.789; Ncor=0.574; logoDP=0.939; NIcor=-0.161; NsEucl=0.874; SSD=2.044; NSW=0.872; rcor=2; rNcor=1; rlogoDP=6; rNIcor=7; rNsEucl=2; rSSD=5; rNSW=2; rank_mean=3.571; match_rank=4
a 0 0 14 4 5 6 0 0 20 2 0
c 0 0 3 0 2 1 0 2 0 0 0
g 0 0 2 12 10 13 16 0 0 9 0
t 0 0 1 4 3 0 4 18 0 9 0
|
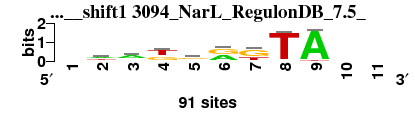
| 3094_NarL_RegulonDB_7.5__shift1 (3094_NarL_RegulonDB_7.5_) |
 |
0.764 |
0.555 |
4.299 |
0.537 |
0.866 |
2.289 |
0.857 |
7 |
3 |
3 |
2 |
4 |
7 |
4 |
4.286 |
5 |
; oligos_7nt_mkv2 versus 3094_NarL_RegulonDB_7.5_; m=5/7; ncol2=8; w=8; offset=1; strand=D; shift=1; score= 4.2857; -wakrrkTA--
; cor=0.764; Ncor=0.555; logoDP=4.299; NIcor=0.537; NsEucl=0.866; SSD=2.289; NSW=0.857; rcor=7; rNcor=3; rlogoDP=3; rNIcor=2; rNsEucl=4; rSSD=7; rNSW=4; rank_mean=4.286; match_rank=5
a 0 38 54 8 25 41 9 0 86 0 0
c 0 4 9 0 12 5 0 4 0 0 0
g 0 17 15 36 45 45 54 2 0 0 0
t 0 32 13 47 9 0 28 85 5 0 0
|
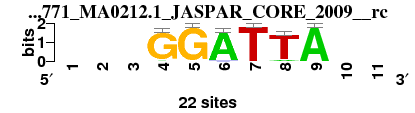
| 1771_MA0212.1_JASPAR_CORE_2009__rc_shift3 (1771_MA0212.1_JASPAR_CORE_2009__rc) |
 |
0.767 |
0.418 |
1.998 |
-0.055 |
0.832 |
2.025 |
0.831 |
5 |
6 |
5 |
5 |
6 |
3 |
6 |
5.143 |
6 |
; oligos_7nt_mkv2 versus 1771_MA0212.1_JASPAR_CORE_2009__rc; m=6/7; ncol2=6; w=6; offset=3; strand=R; shift=3; score= 5.1429; ---GGATTA--
; cor=0.767; Ncor=0.418; logoDP=1.998; NIcor=-0.055; NsEucl=0.832; SSD=2.025; NSW=0.831; rcor=5; rNcor=6; rlogoDP=5; rNIcor=5; rNsEucl=6; rSSD=3; rNSW=6; rank_mean=5.143; match_rank=6
a 0 0 0 1 0 21 0 2 22 0 0
c 0 0 0 0 0 1 0 0 0 0 0
g 0 0 0 21 22 0 0 0 0 0 0
t 0 0 0 0 0 0 22 20 0 0 0
|
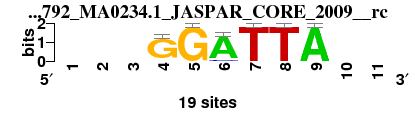
| 1792_MA0234.1_JASPAR_CORE_2009__rc_shift3 (1792_MA0234.1_JASPAR_CORE_2009__rc) |
 |
0.764 |
0.417 |
2.075 |
-0.055 |
0.832 |
2.039 |
0.830 |
6 |
7 |
4 |
4 |
7 |
4 |
7 |
5.571 |
7 |
; oligos_7nt_mkv2 versus 1792_MA0234.1_JASPAR_CORE_2009__rc; m=7/7; ncol2=6; w=6; offset=3; strand=R; shift=3; score= 5.5714; ---GGATTA--
; cor=0.764; Ncor=0.417; logoDP=2.075; NIcor=-0.055; NsEucl=0.832; SSD=2.039; NSW=0.830; rcor=6; rNcor=7; rlogoDP=4; rNIcor=4; rNsEucl=7; rSSD=4; rNSW=7; rank_mean=5.571; match_rank=7
a 0 0 0 1 0 17 0 0 19 0 0
c 0 0 0 1 0 2 0 0 0 0 0
g 0 0 0 17 19 0 0 0 0 0 0
t 0 0 0 0 0 0 19 19 0 0 0
|







